Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAGUGCU
Z-value: 0.27
miRNA associated with seed AAGUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302a-3p
|
MIMAT0000684 |
|
hsa-miR-302b-3p
|
MIMAT0000715 |
|
hsa-miR-302c-3p.1
|
MIMAT0000717 |
|
hsa-miR-302d-3p
|
MIMAT0000718 |
|
hsa-miR-302e
|
MIMAT0005931 |
|
hsa-miR-372-3p
|
MIMAT0000724 |
|
hsa-miR-373-3p
|
MIMAT0000726 |
|
hsa-miR-520a-3p
|
MIMAT0002834 |
|
hsa-miR-520b
|
MIMAT0002843 |
|
hsa-miR-520c-3p
|
MIMAT0002846 |
|
hsa-miR-520d-3p
|
MIMAT0002856 |
|
hsa-miR-520e
|
MIMAT0002825 |
Activity profile of AAGUGCU motif
Sorted Z-values of AAGUGCU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_78526942 | 0.09 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr18_-_29264669 | 0.08 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr8_-_67525473 | 0.08 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr22_+_33197683 | 0.07 |
ENST00000266085.6
|
TIMP3
|
TIMP metallopeptidase inhibitor 3 |
| chr1_-_226076843 | 0.07 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr10_+_65281123 | 0.07 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr18_-_51751132 | 0.07 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr2_-_96811170 | 0.07 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr3_+_140950612 | 0.07 |
ENST00000286353.4
ENST00000502783.1 ENST00000393010.2 ENST00000514680.1 |
ACPL2
|
acid phosphatase-like 2 |
| chr10_+_126490354 | 0.06 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr6_-_108395907 | 0.06 |
ENST00000193322.3
|
OSTM1
|
osteopetrosis associated transmembrane protein 1 |
| chr1_-_207224307 | 0.06 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr4_+_57774042 | 0.06 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr10_-_112064665 | 0.06 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr3_+_172468472 | 0.06 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr4_+_99916765 | 0.05 |
ENST00000296411.6
|
METAP1
|
methionyl aminopeptidase 1 |
| chr17_-_66287257 | 0.05 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr6_+_126112001 | 0.05 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_+_226411319 | 0.05 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr2_-_157189180 | 0.05 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr5_-_56247935 | 0.05 |
ENST00000381199.3
ENST00000381226.3 ENST00000381213.3 |
MIER3
|
mesoderm induction early response 1, family member 3 |
| chr18_-_45456930 | 0.05 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chrX_-_15353629 | 0.05 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr20_+_56884752 | 0.05 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr1_-_85156216 | 0.05 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_-_225450013 | 0.05 |
ENST00000264414.4
|
CUL3
|
cullin 3 |
| chr9_-_16870704 | 0.05 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr4_+_74606223 | 0.05 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr2_-_11484710 | 0.05 |
ENST00000315872.6
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2 |
| chr14_-_45431091 | 0.05 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr11_+_75526212 | 0.05 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr8_+_1772132 | 0.05 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr4_-_76439596 | 0.05 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr4_+_48343339 | 0.05 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr10_+_92980517 | 0.05 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr5_+_98104978 | 0.05 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chr2_+_70056762 | 0.05 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr15_-_25684110 | 0.04 |
ENST00000232165.3
|
UBE3A
|
ubiquitin protein ligase E3A |
| chr2_+_103236004 | 0.04 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr1_+_210001309 | 0.04 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr5_+_133706865 | 0.04 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr5_-_133968529 | 0.04 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr5_+_34656331 | 0.04 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr11_+_33278811 | 0.04 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr16_-_53537105 | 0.04 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr5_-_114880533 | 0.04 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr5_-_114515734 | 0.04 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr16_+_9185450 | 0.04 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr15_-_65809581 | 0.04 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr1_-_20812690 | 0.04 |
ENST00000375078.3
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr2_+_70142189 | 0.04 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr8_-_119634141 | 0.04 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr13_-_107187462 | 0.04 |
ENST00000245323.4
|
EFNB2
|
ephrin-B2 |
| chrX_+_37208521 | 0.04 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr19_-_31840438 | 0.04 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr10_+_70320413 | 0.04 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr17_+_7184986 | 0.04 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr19_+_16222439 | 0.04 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr11_-_64739358 | 0.04 |
ENST00000301896.5
ENST00000530444.1 |
C11orf85
|
chromosome 11 open reading frame 85 |
| chr11_-_102323489 | 0.04 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr2_+_46926048 | 0.04 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr10_+_180987 | 0.04 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr12_-_44200052 | 0.04 |
ENST00000548315.1
ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr1_-_53793584 | 0.04 |
ENST00000354412.3
ENST00000347547.2 ENST00000306052.6 |
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr6_-_89827720 | 0.04 |
ENST00000452027.2
|
SRSF12
|
serine/arginine-rich splicing factor 12 |
| chr15_+_66161871 | 0.04 |
ENST00000569896.1
|
RAB11A
|
RAB11A, member RAS oncogene family |
| chr1_-_197169672 | 0.04 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chrX_+_16737718 | 0.04 |
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1 |
| chr3_-_182698381 | 0.04 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr6_+_43737939 | 0.04 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr13_-_21635631 | 0.04 |
ENST00000382592.4
|
LATS2
|
large tumor suppressor kinase 2 |
| chr6_-_90062543 | 0.03 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr11_-_69519410 | 0.03 |
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19 |
| chrX_+_119384607 | 0.03 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr17_+_30264014 | 0.03 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr16_-_47007545 | 0.03 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr12_-_96184533 | 0.03 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr3_-_149093499 | 0.03 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr14_-_53619816 | 0.03 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr10_-_91295304 | 0.03 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr6_+_158244223 | 0.03 |
ENST00000392185.3
|
SNX9
|
sorting nexin 9 |
| chr2_-_151344172 | 0.03 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr4_-_185395672 | 0.03 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr15_+_68871308 | 0.03 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr17_+_80477571 | 0.03 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr2_-_197036289 | 0.03 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr21_+_47063590 | 0.03 |
ENST00000400314.1
|
PCBP3
|
poly(rC) binding protein 3 |
| chr9_+_33817461 | 0.03 |
ENST00000263228.3
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr8_-_134584152 | 0.03 |
ENST00000521180.1
ENST00000517668.1 ENST00000319914.5 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_-_77532050 | 0.03 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr16_-_3493528 | 0.03 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr1_-_16482554 | 0.03 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr8_+_38854418 | 0.03 |
ENST00000481513.1
ENST00000487273.2 |
ADAM9
|
ADAM metallopeptidase domain 9 |
| chr10_-_123687497 | 0.03 |
ENST00000369040.3
ENST00000224652.6 ENST00000369043.3 |
ATE1
|
arginyltransferase 1 |
| chr3_+_179065474 | 0.03 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr13_-_25746416 | 0.03 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr17_+_38333263 | 0.03 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr13_+_49550015 | 0.03 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr5_+_89770696 | 0.03 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr4_+_44680429 | 0.03 |
ENST00000281543.5
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae) |
| chr9_+_129567282 | 0.03 |
ENST00000449886.1
ENST00000373464.4 ENST00000450858.1 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr3_+_137906109 | 0.03 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr14_-_92506371 | 0.03 |
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr4_-_16228120 | 0.03 |
ENST00000405303.2
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr12_+_45609893 | 0.03 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chr11_+_32851487 | 0.03 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr12_+_27396901 | 0.03 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr1_-_70671216 | 0.03 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr6_+_45389893 | 0.03 |
ENST00000371432.3
|
RUNX2
|
runt-related transcription factor 2 |
| chr13_+_95254085 | 0.03 |
ENST00000376958.4
|
GPR180
|
G protein-coupled receptor 180 |
| chr6_+_149887377 | 0.02 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr1_+_210406121 | 0.02 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr9_-_111929560 | 0.02 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr7_+_6144514 | 0.02 |
ENST00000306177.5
ENST00000465073.2 |
USP42
|
ubiquitin specific peptidase 42 |
| chr2_+_174219548 | 0.02 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chr3_+_43328004 | 0.02 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr1_+_24069642 | 0.02 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chrX_+_14891522 | 0.02 |
ENST00000380492.3
ENST00000482354.1 |
MOSPD2
|
motile sperm domain containing 2 |
| chr5_-_82373260 | 0.02 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr3_+_152017181 | 0.02 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr8_+_26240414 | 0.02 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_-_41909496 | 0.02 |
ENST00000265713.2
ENST00000406337.1 ENST00000396930.3 ENST00000485568.1 ENST00000426524.1 |
KAT6A
|
K(lysine) acetyltransferase 6A |
| chr6_+_108487245 | 0.02 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr19_-_17186229 | 0.02 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr9_+_115513003 | 0.02 |
ENST00000374232.3
|
SNX30
|
sorting nexin family member 30 |
| chr6_-_116381918 | 0.02 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr8_+_28351707 | 0.02 |
ENST00000537916.1
ENST00000523546.1 ENST00000240093.3 |
FZD3
|
frizzled family receptor 3 |
| chr21_-_27542972 | 0.02 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr1_-_205601064 | 0.02 |
ENST00000357992.4
ENST00000289703.4 |
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1) |
| chr10_+_14920843 | 0.02 |
ENST00000433779.1
ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr17_+_5185552 | 0.02 |
ENST00000262477.6
ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr8_-_101965146 | 0.02 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chrX_-_24045303 | 0.02 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr7_+_70597109 | 0.02 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr1_-_235491462 | 0.02 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr17_-_40306934 | 0.02 |
ENST00000592574.1
ENST00000550406.1 ENST00000547517.1 ENST00000393860.3 ENST00000346213.4 |
CTD-2132N18.3
RAB5C
|
Uncharacterized protein RAB5C, member RAS oncogene family |
| chr14_-_77965151 | 0.02 |
ENST00000393684.3
ENST00000493585.1 ENST00000554801.2 ENST00000342219.4 ENST00000412904.1 ENST00000429906.1 |
ISM2
|
isthmin 2 |
| chr1_+_89149905 | 0.02 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr15_+_38544476 | 0.02 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr8_-_37756972 | 0.02 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr1_+_162039558 | 0.02 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr4_+_156588350 | 0.02 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr13_-_29069232 | 0.02 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr3_-_69435224 | 0.02 |
ENST00000398540.3
|
FRMD4B
|
FERM domain containing 4B |
| chr3_+_119013185 | 0.02 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr2_-_68479614 | 0.02 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr2_+_179345173 | 0.02 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr5_-_168006591 | 0.02 |
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3 |
| chr14_+_36295504 | 0.02 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr10_+_71078595 | 0.02 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr5_+_170814803 | 0.02 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr6_-_90121938 | 0.02 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr16_-_77468945 | 0.02 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr7_+_139026057 | 0.02 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr9_-_99180597 | 0.02 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr18_+_42260861 | 0.02 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr9_+_128509624 | 0.02 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr3_-_48229846 | 0.02 |
ENST00000302506.3
ENST00000351231.3 ENST00000437972.1 |
CDC25A
|
cell division cycle 25A |
| chr18_-_19284724 | 0.02 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr21_-_36260980 | 0.02 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr3_-_171178157 | 0.02 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr6_+_87865262 | 0.02 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr12_+_72148614 | 0.02 |
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family |
| chr13_+_97874574 | 0.02 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_-_99395787 | 0.02 |
ENST00000369244.2
ENST00000229971.1 |
FBXL4
|
F-box and leucine-rich repeat protein 4 |
| chr6_+_18155560 | 0.02 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr5_+_56111361 | 0.02 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr18_-_33709268 | 0.02 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr7_-_75368248 | 0.02 |
ENST00000434438.2
ENST00000336926.6 |
HIP1
|
huntingtin interacting protein 1 |
| chr4_+_128703295 | 0.02 |
ENST00000296464.4
ENST00000508549.1 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr17_+_73717516 | 0.02 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr4_-_83719983 | 0.02 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr4_-_111119804 | 0.02 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_111967345 | 0.02 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr5_+_112043186 | 0.02 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr17_+_38278530 | 0.02 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr1_-_161039456 | 0.02 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr12_-_77459306 | 0.02 |
ENST00000547316.1
ENST00000416496.2 ENST00000550669.1 ENST00000322886.7 |
E2F7
|
E2F transcription factor 7 |
| chr1_+_3607228 | 0.02 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr5_+_74807581 | 0.02 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr3_+_20081515 | 0.02 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr12_-_56583332 | 0.02 |
ENST00000347471.4
ENST00000267064.4 ENST00000394023.3 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr11_+_9406169 | 0.02 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr8_-_53626974 | 0.02 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr5_-_1882858 | 0.02 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr1_-_94703118 | 0.02 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr20_-_656823 | 0.02 |
ENST00000246104.6
|
SCRT2
|
scratch family zinc finger 2 |
| chr5_-_174871136 | 0.02 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr3_+_196466710 | 0.02 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr4_+_88928777 | 0.02 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr12_+_120875910 | 0.02 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr10_+_97803151 | 0.02 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr12_+_83080659 | 0.02 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr14_+_67826709 | 0.02 |
ENST00000256383.4
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr5_+_63461642 | 0.02 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr1_-_55680762 | 0.02 |
ENST00000407756.1
ENST00000294383.6 |
USP24
|
ubiquitin specific peptidase 24 |
| chr2_-_25194963 | 0.02 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr5_+_172483347 | 0.02 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chr5_+_149109825 | 0.02 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr13_-_26795840 | 0.02 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
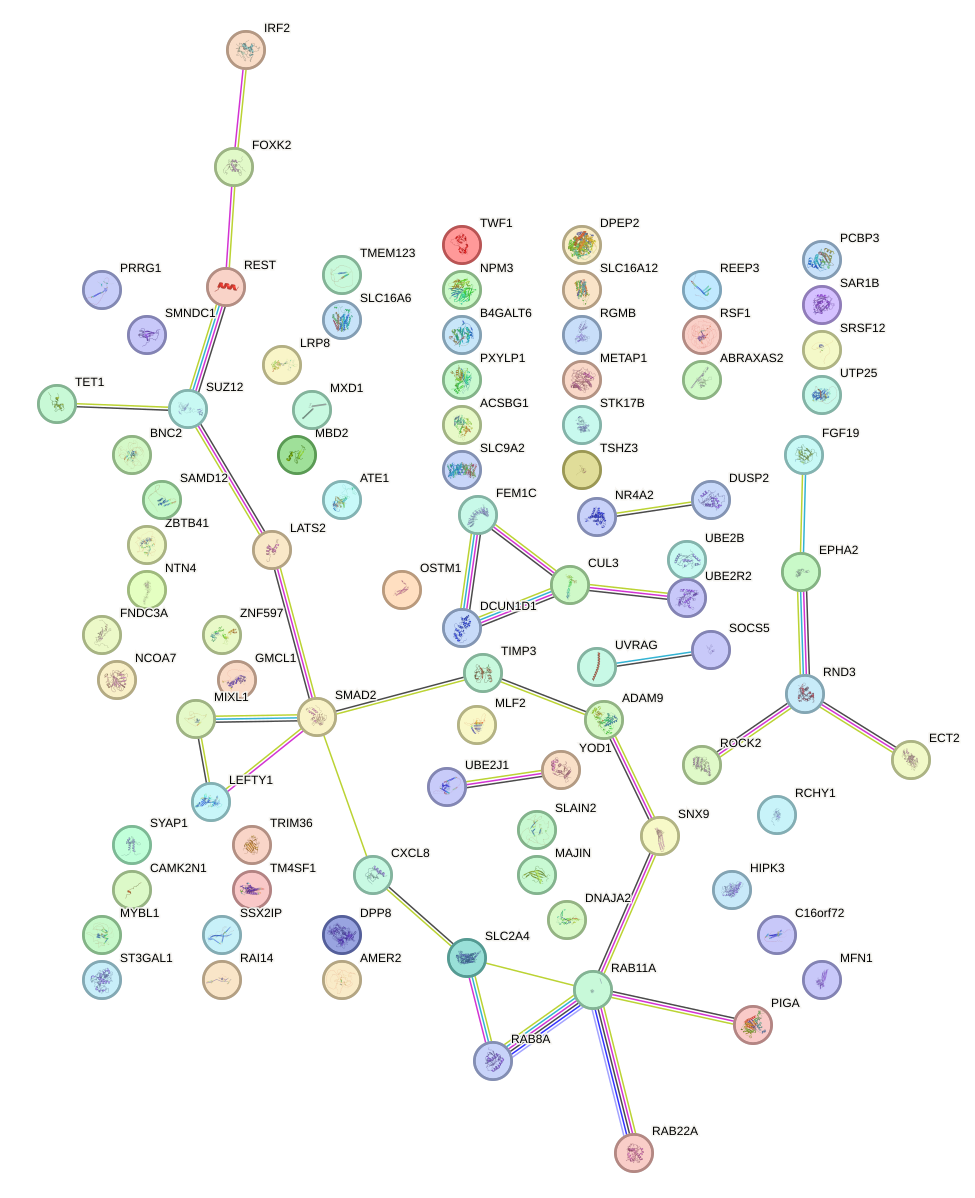
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGUGCU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |