Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AHR_ARNT2
Z-value: 0.48
Transcription factors associated with AHR_ARNT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AHR
|
ENSG00000106546.8 | aryl hydrocarbon receptor |
|
ARNT2
|
ENSG00000172379.14 | aryl hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT2 | hg19_v2_chr15_+_80696666_80696700 | 0.53 | 4.7e-01 | Click! |
| AHR | hg19_v2_chr7_+_17338239_17338262 | -0.15 | 8.5e-01 | Click! |
Activity profile of AHR_ARNT2 motif
Sorted Z-values of AHR_ARNT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_163834852 | 0.37 |
ENST00000604200.1
|
CAHM
|
colon adenocarcinoma hypermethylated (non-protein coding) |
| chr5_-_81046904 | 0.26 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr14_-_23791484 | 0.25 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr11_-_568369 | 0.23 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chr12_+_77158021 | 0.23 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chrX_-_70288234 | 0.22 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr16_+_30709530 | 0.22 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr11_-_65314905 | 0.20 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr10_+_104178946 | 0.20 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr16_-_4664382 | 0.20 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr3_+_49044765 | 0.19 |
ENST00000429900.2
|
WDR6
|
WD repeat domain 6 |
| chr2_+_71295766 | 0.19 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr14_-_21852119 | 0.18 |
ENST00000555943.1
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae) |
| chr16_-_28223229 | 0.18 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr4_-_1107306 | 0.18 |
ENST00000433731.2
ENST00000333673.5 ENST00000382968.5 |
RNF212
|
ring finger protein 212 |
| chr12_+_49761147 | 0.17 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr19_-_46145696 | 0.17 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr1_+_156308245 | 0.17 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr2_-_230787879 | 0.17 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr12_-_9102224 | 0.16 |
ENST00000543845.1
ENST00000544245.1 |
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr22_+_46972975 | 0.15 |
ENST00000431155.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr19_-_663147 | 0.15 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr16_+_3019309 | 0.15 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr3_-_171177852 | 0.15 |
ENST00000284483.8
ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr5_-_81046841 | 0.15 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr12_+_9102632 | 0.14 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr19_-_44123734 | 0.14 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr22_-_39548627 | 0.14 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr3_-_156534754 | 0.14 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr15_+_42565393 | 0.13 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr7_-_105926058 | 0.13 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr12_+_112451222 | 0.13 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr3_-_135915146 | 0.12 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr17_+_79369249 | 0.12 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr9_-_14322319 | 0.12 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr6_-_38607628 | 0.12 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr1_+_109642799 | 0.12 |
ENST00000602755.1
|
SCARNA2
|
small Cajal body-specific RNA 2 |
| chr11_+_125439176 | 0.12 |
ENST00000529812.1
|
EI24
|
etoposide induced 2.4 |
| chr5_-_100238918 | 0.12 |
ENST00000451528.2
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr6_-_10747802 | 0.12 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr3_+_157261116 | 0.12 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr10_-_38146965 | 0.12 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr14_+_102414651 | 0.12 |
ENST00000607414.1
|
RP11-1017G21.5
|
RP11-1017G21.5 |
| chr19_+_18718214 | 0.11 |
ENST00000600490.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr12_+_111856144 | 0.11 |
ENST00000550925.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr22_-_39548443 | 0.11 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr20_-_1373606 | 0.11 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr22_-_39151995 | 0.11 |
ENST00000405018.1
ENST00000438058.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr2_+_232575168 | 0.11 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr16_-_54963026 | 0.11 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_-_762885 | 0.11 |
ENST00000536430.1
ENST00000473798.1 |
LINC00115
|
long intergenic non-protein coding RNA 115 |
| chr5_-_176730733 | 0.10 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr12_-_80084333 | 0.10 |
ENST00000552637.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr2_-_24346218 | 0.10 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr2_-_175202151 | 0.10 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr17_-_58603482 | 0.10 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr19_+_49956426 | 0.10 |
ENST00000293350.4
ENST00000540132.1 ENST00000455361.2 ENST00000433981.2 |
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr8_+_120885949 | 0.10 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr1_+_156308403 | 0.10 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr2_+_203499901 | 0.10 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr9_+_127539425 | 0.10 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr5_+_149865377 | 0.09 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr1_+_45140360 | 0.09 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr10_+_111985837 | 0.09 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr15_+_41952591 | 0.09 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr19_-_5624057 | 0.09 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr15_-_72410455 | 0.09 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr2_+_71295733 | 0.09 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr11_-_116968987 | 0.09 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr3_-_42003613 | 0.09 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr22_-_39548511 | 0.09 |
ENST00000434260.1
|
CBX7
|
chromobox homolog 7 |
| chr19_-_59010565 | 0.09 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr5_+_32712363 | 0.09 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr19_-_49140692 | 0.09 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_-_64062583 | 0.09 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chr19_+_5681153 | 0.09 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr19_-_30205963 | 0.09 |
ENST00000392278.2
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr17_-_73178599 | 0.09 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr1_+_113392455 | 0.09 |
ENST00000456651.1
ENST00000422022.1 |
RP3-522D1.1
|
RP3-522D1.1 |
| chr15_+_42565464 | 0.09 |
ENST00000562170.1
ENST00000562859.1 |
GANC
|
glucosidase, alpha; neutral C |
| chr2_+_30670209 | 0.08 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr3_+_112930946 | 0.08 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr19_-_49140609 | 0.08 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr1_-_8939265 | 0.08 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr7_-_155089251 | 0.08 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr7_+_150748288 | 0.08 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr20_-_1165319 | 0.08 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr1_-_1535455 | 0.08 |
ENST00000422725.1
|
C1orf233
|
chromosome 1 open reading frame 233 |
| chr20_-_1165117 | 0.08 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr7_+_12250943 | 0.08 |
ENST00000442107.1
|
TMEM106B
|
transmembrane protein 106B |
| chr20_-_3219359 | 0.08 |
ENST00000437836.2
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr5_-_38557561 | 0.08 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr8_+_110552046 | 0.08 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr19_+_18530184 | 0.08 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr9_+_37079968 | 0.08 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr22_-_39151947 | 0.08 |
ENST00000216064.4
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr1_+_45477901 | 0.08 |
ENST00000434478.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr15_-_75932528 | 0.08 |
ENST00000403490.1
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr19_+_46144884 | 0.08 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chrX_-_128788914 | 0.08 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr22_-_50765489 | 0.08 |
ENST00000413817.3
|
DENND6B
|
DENN/MADD domain containing 6B |
| chr17_-_8059638 | 0.08 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr12_+_32259819 | 0.08 |
ENST00000550207.1
|
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr16_-_1823114 | 0.08 |
ENST00000177742.3
ENST00000397375.2 |
MRPS34
|
mitochondrial ribosomal protein S34 |
| chr19_-_55672037 | 0.08 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr16_+_58497587 | 0.08 |
ENST00000569404.1
ENST00000569539.1 ENST00000564126.1 ENST00000565304.1 ENST00000567667.1 |
NDRG4
|
NDRG family member 4 |
| chr1_+_6845384 | 0.08 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr4_-_83351294 | 0.08 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr15_-_45670717 | 0.08 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_+_3019246 | 0.08 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr22_+_29664241 | 0.07 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr8_+_142402089 | 0.07 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr19_-_47164386 | 0.07 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr20_+_19738792 | 0.07 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr3_-_142720267 | 0.07 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr12_+_82752283 | 0.07 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr1_+_197871854 | 0.07 |
ENST00000436652.1
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr19_-_10213335 | 0.07 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr19_-_14201776 | 0.07 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr5_+_134368954 | 0.07 |
ENST00000432382.3
|
CTC-349C3.1
|
chromosome 5 open reading frame 66 |
| chr8_-_101964738 | 0.07 |
ENST00000523938.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chrX_-_83442915 | 0.07 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr9_-_138391692 | 0.07 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr22_-_29663690 | 0.07 |
ENST00000406335.1
|
RHBDD3
|
rhomboid domain containing 3 |
| chr12_-_123717711 | 0.07 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr1_-_1208851 | 0.07 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr9_-_139760717 | 0.07 |
ENST00000371648.4
|
EDF1
|
endothelial differentiation-related factor 1 |
| chr1_+_44870866 | 0.07 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr3_+_52570610 | 0.07 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr1_-_91317072 | 0.07 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr19_+_18529674 | 0.07 |
ENST00000597724.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr10_-_977564 | 0.07 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr1_+_92495528 | 0.07 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr4_+_128886584 | 0.07 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr10_+_118608998 | 0.06 |
ENST00000409522.1
ENST00000341276.5 ENST00000512864.2 |
ENO4
|
enolase family member 4 |
| chr20_+_34680595 | 0.06 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr5_-_81046922 | 0.06 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr15_+_41913690 | 0.06 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr12_+_57482665 | 0.06 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr19_-_44258770 | 0.06 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_-_2427536 | 0.06 |
ENST00000591871.1
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr6_-_10412600 | 0.06 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr7_-_86849836 | 0.06 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr4_+_170541678 | 0.06 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr16_-_8962853 | 0.06 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr14_+_105559784 | 0.06 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr8_+_42396936 | 0.06 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr4_+_130014836 | 0.06 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr1_+_61547894 | 0.06 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr9_-_139760737 | 0.06 |
ENST00000371649.1
ENST00000224073.1 |
EDF1
|
endothelial differentiation-related factor 1 |
| chr10_+_114710516 | 0.06 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_72057571 | 0.06 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr19_-_44124019 | 0.06 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr16_+_2059872 | 0.06 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr10_+_104404644 | 0.06 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr5_+_174905532 | 0.06 |
ENST00000502393.1
ENST00000506963.1 |
SFXN1
|
sideroflexin 1 |
| chr7_+_74072288 | 0.06 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr15_+_42565844 | 0.06 |
ENST00000566442.1
|
GANC
|
glucosidase, alpha; neutral C |
| chrX_-_30327495 | 0.06 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr16_-_67217844 | 0.06 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr5_-_36241900 | 0.06 |
ENST00000381937.4
ENST00000514504.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr11_-_87908600 | 0.06 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr5_+_56205878 | 0.06 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr4_+_183370146 | 0.06 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr2_+_177502438 | 0.06 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr2_+_149402009 | 0.06 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_+_150565038 | 0.06 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chrX_+_134555863 | 0.06 |
ENST00000417443.2
|
LINC00086
|
long intergenic non-protein coding RNA 86 |
| chr3_-_171178157 | 0.06 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr6_-_30043539 | 0.06 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr1_+_234509186 | 0.06 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr8_+_38854368 | 0.06 |
ENST00000466936.1
|
ADAM9
|
ADAM metallopeptidase domain 9 |
| chr19_+_54606145 | 0.06 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr12_+_122356488 | 0.06 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr3_+_138067521 | 0.06 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr1_-_85930246 | 0.06 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_695787 | 0.06 |
ENST00000526170.1
ENST00000488769.1 |
TMEM80
|
transmembrane protein 80 |
| chr22_-_37823468 | 0.06 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr1_-_109203685 | 0.06 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr7_+_45613958 | 0.06 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr4_-_129209221 | 0.06 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr5_+_149340339 | 0.06 |
ENST00000433184.1
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr17_+_41177220 | 0.06 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr14_+_64970662 | 0.06 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr12_-_53614043 | 0.06 |
ENST00000338561.5
|
RARG
|
retinoic acid receptor, gamma |
| chr11_+_61447845 | 0.06 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr6_-_27841289 | 0.06 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr16_-_88729473 | 0.06 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr14_-_23299009 | 0.06 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_52859046 | 0.06 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_+_37458904 | 0.06 |
ENST00000416653.1
|
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr3_+_197477038 | 0.06 |
ENST00000426031.1
ENST00000424384.2 |
FYTTD1
|
forty-two-three domain containing 1 |
| chr10_-_46342675 | 0.06 |
ENST00000492347.1
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr8_-_42751728 | 0.06 |
ENST00000319104.3
ENST00000531440.1 |
RNF170
|
ring finger protein 170 |
| chr19_-_54974894 | 0.06 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr15_-_32162833 | 0.06 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr6_+_29691056 | 0.05 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr5_+_139781393 | 0.05 |
ENST00000360839.2
ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr16_-_67969888 | 0.05 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr4_-_83350580 | 0.05 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
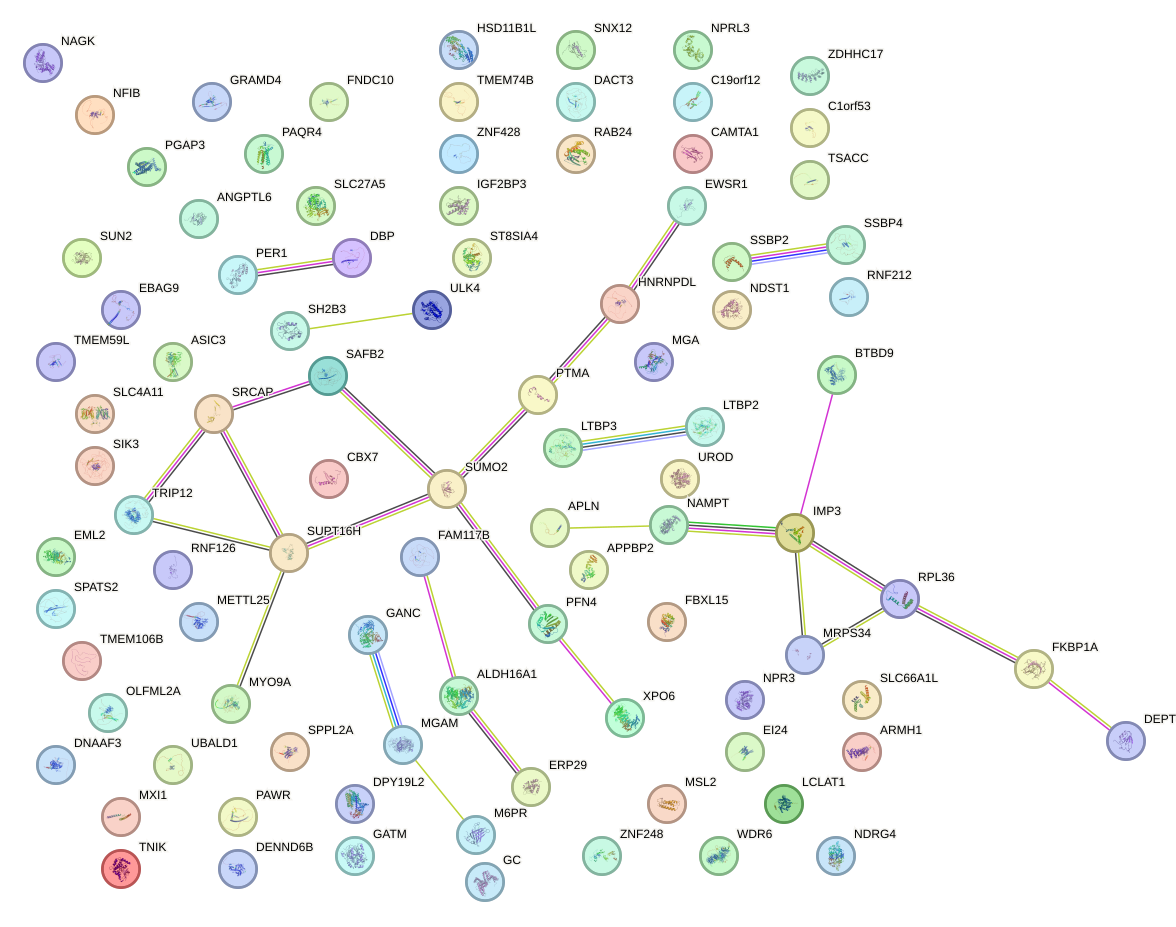
Network of associatons between targets according to the STRING database.
First level regulatory network of AHR_ARNT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.2 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.4 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.2 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:0072080 | nephron tubule development(GO:0072080) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0021622 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.0 | GO:0048242 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) negative regulation of luteinizing hormone secretion(GO:0033685) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.0 | GO:0010498 | proteasomal protein catabolic process(GO:0010498) proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.0 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.0 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.4 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.0 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |