Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CCACAGG
Z-value: 0.32
miRNA associated with seed CCACAGG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-140-3p.1
|
Activity profile of CCACAGG motif
Sorted Z-values of CCACAGG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_56652111 | 0.06 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr12_+_74931551 | 0.06 |
ENST00000519948.2
|
ATXN7L3B
|
ataxin 7-like 3B |
| chr1_-_228135599 | 0.05 |
ENST00000272164.5
|
WNT9A
|
wingless-type MMTV integration site family, member 9A |
| chr22_+_30115986 | 0.05 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr11_+_118754475 | 0.05 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr19_-_47616992 | 0.05 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr9_-_127533519 | 0.05 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr12_+_57916466 | 0.05 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr17_+_42634844 | 0.05 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr9_-_131592058 | 0.04 |
ENST00000361256.5
|
C9orf114
|
chromosome 9 open reading frame 114 |
| chr5_+_149887672 | 0.04 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_-_81787006 | 0.04 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr16_+_3019246 | 0.04 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr14_+_23790655 | 0.04 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr15_+_40763150 | 0.04 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr1_+_19970657 | 0.04 |
ENST00000375136.3
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr19_-_44100275 | 0.04 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr3_-_48632593 | 0.04 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr5_-_180242534 | 0.03 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr16_+_28303804 | 0.03 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr1_-_21671968 | 0.03 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr3_-_114790179 | 0.03 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr22_-_38669030 | 0.03 |
ENST00000361906.3
|
TMEM184B
|
transmembrane protein 184B |
| chr17_-_43568062 | 0.03 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr17_+_61699766 | 0.03 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr19_+_55738587 | 0.03 |
ENST00000598855.1
|
AC010327.2
|
Uncharacterized protein; cDNA FLJ45856 fis, clone OCBBF2025631 |
| chr20_+_306221 | 0.03 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr17_-_79885576 | 0.03 |
ENST00000574686.1
ENST00000357736.4 |
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr5_-_137090028 | 0.03 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr2_+_26987111 | 0.03 |
ENST00000344420.5
ENST00000416475.2 |
SLC35F6
|
solute carrier family 35, member F6 |
| chr17_+_42422637 | 0.03 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr1_+_27153173 | 0.03 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr17_+_73452545 | 0.03 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr17_-_3571934 | 0.03 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr10_-_126432619 | 0.03 |
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B |
| chr1_+_36348790 | 0.03 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr17_-_33446820 | 0.03 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr17_+_7452189 | 0.03 |
ENST00000293825.6
|
TNFSF12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr20_+_44462430 | 0.03 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr14_+_65171099 | 0.03 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_-_75062730 | 0.02 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr16_+_31085714 | 0.02 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr11_-_60929074 | 0.02 |
ENST00000301765.5
|
VPS37C
|
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr19_+_41816053 | 0.02 |
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97 |
| chr15_+_40733387 | 0.02 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr20_+_62526467 | 0.02 |
ENST00000369911.2
ENST00000360864.4 |
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5 |
| chr17_+_60704762 | 0.02 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr5_+_140044261 | 0.02 |
ENST00000358337.5
|
WDR55
|
WD repeat domain 55 |
| chr22_-_42017021 | 0.02 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr7_+_100136811 | 0.02 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr16_-_85833160 | 0.02 |
ENST00000435200.2
|
EMC8
|
ER membrane protein complex subunit 8 |
| chr15_+_41851211 | 0.02 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr14_-_77279153 | 0.02 |
ENST00000251089.2
|
ANGEL1
|
angel homolog 1 (Drosophila) |
| chr1_-_154943212 | 0.02 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr14_-_20929624 | 0.02 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chrX_-_153237258 | 0.02 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr6_-_34664612 | 0.02 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr1_+_113010056 | 0.02 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr20_-_50384864 | 0.02 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr9_+_133710453 | 0.02 |
ENST00000318560.5
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr11_+_12695944 | 0.02 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr19_+_38397839 | 0.02 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr5_-_175964366 | 0.02 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr19_-_14316980 | 0.02 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr3_-_44519131 | 0.02 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr19_-_49140692 | 0.02 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr5_+_154092396 | 0.02 |
ENST00000336314.4
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr17_-_4167142 | 0.02 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr11_-_70507901 | 0.02 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr22_+_51112800 | 0.02 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_+_43855560 | 0.02 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr15_+_89631381 | 0.02 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr11_+_73087309 | 0.02 |
ENST00000064780.2
ENST00000545687.1 |
RELT
|
RELT tumor necrosis factor receptor |
| chr14_+_70078303 | 0.02 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr17_-_3794021 | 0.02 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr14_-_39901618 | 0.02 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr1_-_26197744 | 0.02 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr1_+_18958008 | 0.02 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr3_-_66551351 | 0.02 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr11_-_62494821 | 0.02 |
ENST00000301785.5
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr15_+_91473403 | 0.02 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr17_-_26733179 | 0.02 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr3_+_51428704 | 0.01 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr17_+_1958388 | 0.01 |
ENST00000399849.3
|
HIC1
|
hypermethylated in cancer 1 |
| chr17_-_42298331 | 0.01 |
ENST00000343638.5
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr17_+_36886478 | 0.01 |
ENST00000439660.2
|
CISD3
|
CDGSH iron sulfur domain 3 |
| chr15_+_89164520 | 0.01 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr17_-_61523535 | 0.01 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr9_+_133884469 | 0.01 |
ENST00000361069.4
|
LAMC3
|
laminin, gamma 3 |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr11_+_119076745 | 0.01 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr3_+_127771212 | 0.01 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr6_-_116601044 | 0.01 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr7_-_135662056 | 0.01 |
ENST00000393085.3
ENST00000435723.1 |
MTPN
|
myotrophin |
| chr19_-_18632861 | 0.01 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr10_+_11206925 | 0.01 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr15_+_80696666 | 0.01 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr5_+_42423872 | 0.01 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr2_-_43453734 | 0.01 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr10_+_57358750 | 0.01 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr2_-_166930131 | 0.01 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr7_+_97736197 | 0.01 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr3_+_14989076 | 0.01 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chrX_-_124097620 | 0.01 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr17_+_47865917 | 0.01 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr3_-_50374869 | 0.01 |
ENST00000327761.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr17_+_6918093 | 0.01 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr12_-_42538657 | 0.01 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr5_-_77844974 | 0.01 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr14_-_77787198 | 0.01 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr17_-_36762095 | 0.01 |
ENST00000578925.1
ENST00000264659.7 |
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr7_+_44084262 | 0.01 |
ENST00000456905.1
ENST00000440166.1 ENST00000452943.1 ENST00000468694.1 ENST00000494774.1 ENST00000490734.2 |
DBNL
|
drebrin-like |
| chr2_-_96931679 | 0.01 |
ENST00000258439.3
ENST00000432959.1 |
TMEM127
|
transmembrane protein 127 |
| chr10_+_63661053 | 0.01 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr5_+_112312416 | 0.01 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr1_-_113249948 | 0.01 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr16_+_2587998 | 0.01 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr16_+_23652700 | 0.01 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr20_-_4982132 | 0.01 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr15_+_91411810 | 0.01 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr18_-_53255766 | 0.01 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr4_+_15004165 | 0.01 |
ENST00000538197.1
ENST00000541112.1 ENST00000442003.2 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr14_+_56585048 | 0.01 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_-_130786400 | 0.01 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr6_+_36164487 | 0.01 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr6_-_62996066 | 0.01 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr14_+_71108460 | 0.01 |
ENST00000256367.2
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr16_+_50775948 | 0.01 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr1_-_16678914 | 0.01 |
ENST00000375592.3
|
FBXO42
|
F-box protein 42 |
| chr11_+_63580855 | 0.01 |
ENST00000294244.4
|
C11orf84
|
chromosome 11 open reading frame 84 |
| chr3_+_49591881 | 0.01 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr1_-_32801825 | 0.01 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr8_-_116681221 | 0.01 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_+_35684288 | 0.01 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr1_-_151300160 | 0.01 |
ENST00000368874.4
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr19_+_59055814 | 0.01 |
ENST00000594806.1
ENST00000253024.5 ENST00000341753.6 |
TRIM28
|
tripartite motif containing 28 |
| chr20_+_1875110 | 0.01 |
ENST00000400068.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr3_+_85008089 | 0.01 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr5_-_178017355 | 0.01 |
ENST00000390654.3
|
COL23A1
|
collagen, type XXIII, alpha 1 |
| chr12_+_122150646 | 0.01 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr9_-_14314066 | 0.01 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr18_+_18943554 | 0.01 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr14_-_68283291 | 0.01 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chr12_-_50101165 | 0.01 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chrX_+_150151752 | 0.01 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr11_-_64901978 | 0.01 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr10_-_75571341 | 0.01 |
ENST00000309979.6
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chrX_-_134186144 | 0.01 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr8_+_77593448 | 0.01 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_+_36322408 | 0.01 |
ENST00000361383.6
ENST00000447935.1 ENST00000405275.2 |
CTNNBL1
|
catenin, beta like 1 |
| chr22_+_29279552 | 0.01 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr10_+_99400443 | 0.01 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr1_+_165796753 | 0.01 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr7_+_140774032 | 0.01 |
ENST00000565468.1
|
TMEM178B
|
transmembrane protein 178B |
| chr3_-_32022733 | 0.01 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr5_+_143584814 | 0.01 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr2_-_54483384 | 0.01 |
ENST00000317802.7
|
TSPYL6
|
TSPY-like 6 |
| chr5_+_32788945 | 0.01 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr19_+_34745442 | 0.01 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chr11_+_48002076 | 0.01 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr16_+_81348528 | 0.01 |
ENST00000568107.2
|
GAN
|
gigaxonin |
| chr1_+_168195229 | 0.01 |
ENST00000271375.4
ENST00000367825.3 |
SFT2D2
|
SFT2 domain containing 2 |
| chr16_+_24741013 | 0.01 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr14_-_74253948 | 0.01 |
ENST00000394071.2
|
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr1_-_209979375 | 0.01 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr17_+_56160768 | 0.01 |
ENST00000579991.2
|
DYNLL2
|
dynein, light chain, LC8-type 2 |
| chr9_+_37753795 | 0.00 |
ENST00000377753.2
ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B
|
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chr17_-_39890893 | 0.00 |
ENST00000393939.2
ENST00000347901.4 ENST00000341193.5 ENST00000310778.5 |
HAP1
|
huntingtin-associated protein 1 |
| chr9_-_100684845 | 0.00 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr9_+_116638562 | 0.00 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr20_-_44718538 | 0.00 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr15_-_37390482 | 0.00 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr2_-_166060571 | 0.00 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr9_-_107690420 | 0.00 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr16_+_11038345 | 0.00 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr3_-_57199397 | 0.00 |
ENST00000296318.7
|
IL17RD
|
interleukin 17 receptor D |
| chr7_+_128864848 | 0.00 |
ENST00000325006.3
ENST00000446544.2 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr17_-_62502639 | 0.00 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_-_151319710 | 0.00 |
ENST00000290524.4
ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr4_+_118955500 | 0.00 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr19_-_14629224 | 0.00 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr12_+_50898881 | 0.00 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr11_-_63439013 | 0.00 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr4_+_145567173 | 0.00 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr8_-_98290087 | 0.00 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr17_-_40897043 | 0.00 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
| chr7_-_132261253 | 0.00 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr1_+_10271674 | 0.00 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr17_+_38296576 | 0.00 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr4_+_134070439 | 0.00 |
ENST00000264360.5
|
PCDH10
|
protocadherin 10 |
| chr3_+_48956249 | 0.00 |
ENST00000452882.1
ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr13_+_52158610 | 0.00 |
ENST00000298125.5
|
WDFY2
|
WD repeat and FYVE domain containing 2 |
| chr2_-_200322723 | 0.00 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr2_+_169312350 | 0.00 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr15_-_42264702 | 0.00 |
ENST00000220325.4
|
EHD4
|
EH-domain containing 4 |
| chr3_-_52312636 | 0.00 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr1_+_16010779 | 0.00 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr5_+_102594403 | 0.00 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr5_-_171433819 | 0.00 |
ENST00000296933.6
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr14_-_77843390 | 0.00 |
ENST00000216468.7
|
TMED8
|
transmembrane emp24 protein transport domain containing 8 |
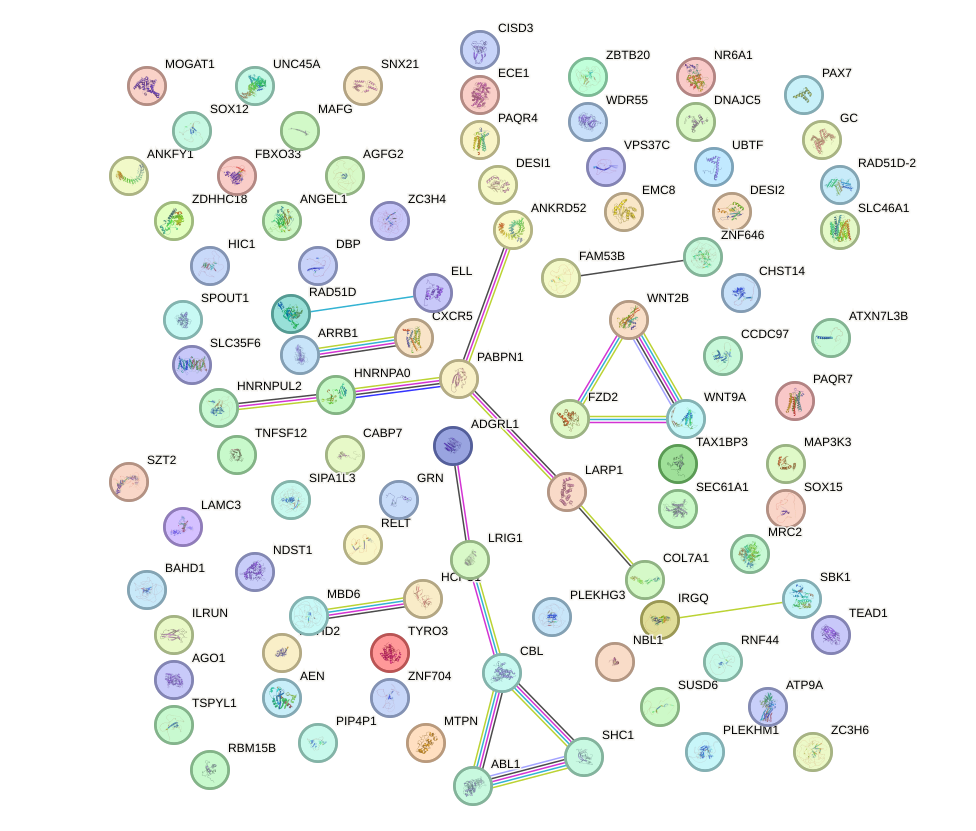
Network of associatons between targets according to the STRING database.
First level regulatory network of CCACAGG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |