Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CDX1
Z-value: 0.41
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | caudal type homeobox 1 |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_62439727 | 0.11 |
ENST00000528862.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr2_+_227771404 | 0.11 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr16_+_2014941 | 0.10 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr16_+_56691606 | 0.09 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr17_-_57229155 | 0.09 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr18_-_68004529 | 0.09 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr1_+_112016414 | 0.09 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr15_+_49715449 | 0.09 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr14_+_39703112 | 0.08 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_+_149447783 | 0.08 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr10_-_69597828 | 0.08 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr9_-_3469181 | 0.08 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr9_-_95244781 | 0.07 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr2_+_132479948 | 0.07 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr3_-_96337000 | 0.07 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr6_-_138866823 | 0.07 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr10_-_18948208 | 0.07 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr21_+_25801041 | 0.07 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr10_-_63995871 | 0.06 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr14_+_88490894 | 0.06 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr17_-_67057047 | 0.06 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr3_-_148939598 | 0.06 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr15_+_84841242 | 0.06 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr17_+_65375082 | 0.06 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_-_54411255 | 0.06 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr5_-_95550754 | 0.06 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr10_-_69597915 | 0.06 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr8_+_76452097 | 0.06 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr2_-_151344172 | 0.06 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr1_-_54411240 | 0.06 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr2_+_192543694 | 0.06 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_-_48943706 | 0.06 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr15_+_49715293 | 0.06 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr13_-_24476794 | 0.06 |
ENST00000382140.2
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr4_+_175205100 | 0.05 |
ENST00000515299.1
|
CEP44
|
centrosomal protein 44kDa |
| chr18_+_47901408 | 0.05 |
ENST00000398452.2
ENST00000417656.2 ENST00000488454.1 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr15_+_55700741 | 0.05 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr2_-_203735484 | 0.05 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr10_-_95360983 | 0.05 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr5_-_79946820 | 0.05 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr16_+_2014993 | 0.05 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr3_+_186285192 | 0.05 |
ENST00000439351.1
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr4_+_124571409 | 0.05 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr18_-_56985873 | 0.05 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr11_-_8795787 | 0.05 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr7_-_92777606 | 0.05 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr7_-_33080506 | 0.05 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr1_+_161736072 | 0.05 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr16_-_28192360 | 0.05 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr11_+_74811578 | 0.05 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr16_-_67965756 | 0.05 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr4_-_52883786 | 0.05 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr6_+_144665237 | 0.05 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr13_-_73301819 | 0.05 |
ENST00000377818.3
|
MZT1
|
mitotic spindle organizing protein 1 |
| chr17_-_46690839 | 0.05 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr11_-_77122928 | 0.05 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr17_+_41005283 | 0.04 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr16_-_1275257 | 0.04 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr7_+_141463897 | 0.04 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chrY_+_14958970 | 0.04 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr10_+_127661942 | 0.04 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr12_-_76461249 | 0.04 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_+_112047087 | 0.04 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr1_+_104159999 | 0.04 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr4_+_74606223 | 0.04 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr18_+_20513782 | 0.04 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr1_+_200993071 | 0.04 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr8_+_93895865 | 0.04 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr4_-_100485143 | 0.04 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr3_-_196987309 | 0.04 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr13_+_53030107 | 0.04 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr18_-_29264669 | 0.04 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr17_+_22022437 | 0.04 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr8_-_125551278 | 0.04 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr15_+_45879964 | 0.04 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr12_+_16500037 | 0.04 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_-_36813162 | 0.04 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr5_+_75904918 | 0.04 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr19_+_38880695 | 0.04 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr1_+_150254936 | 0.04 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr9_-_69229650 | 0.04 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr2_+_192543153 | 0.04 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr22_+_18111594 | 0.04 |
ENST00000399782.1
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr10_+_112327425 | 0.04 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr17_-_57232596 | 0.04 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_+_97306295 | 0.04 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr5_+_118691706 | 0.04 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_+_58797974 | 0.04 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr6_-_128222103 | 0.04 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr14_+_56127960 | 0.03 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_+_53196872 | 0.03 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr18_-_29264467 | 0.03 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr15_+_77713299 | 0.03 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr5_+_96079240 | 0.03 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr8_+_16884740 | 0.03 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr14_-_67826538 | 0.03 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr2_-_44550441 | 0.03 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr4_-_83765613 | 0.03 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr11_-_4719072 | 0.03 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr14_+_24407940 | 0.03 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr3_-_49314640 | 0.03 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr8_-_112248400 | 0.03 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr13_-_103346854 | 0.03 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr12_+_32654965 | 0.03 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr20_+_16729003 | 0.03 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr18_+_61575200 | 0.03 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_-_197226875 | 0.03 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_+_68164752 | 0.03 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr15_-_76304731 | 0.03 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr3_+_171561127 | 0.03 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr8_+_66619277 | 0.03 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr11_-_5537920 | 0.03 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr5_+_179135246 | 0.03 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr5_-_133510456 | 0.03 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr3_-_197025447 | 0.03 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr13_+_76334498 | 0.03 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr3_+_190333097 | 0.03 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr14_-_53331239 | 0.03 |
ENST00000553663.1
|
FERMT2
|
fermitin family member 2 |
| chr5_-_150138061 | 0.03 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chrX_-_45629661 | 0.03 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chrX_-_119693745 | 0.03 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr6_+_76599809 | 0.03 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr14_+_102276209 | 0.03 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr16_-_72206034 | 0.03 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr17_-_39023462 | 0.03 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr12_+_27398584 | 0.03 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr5_-_96518907 | 0.03 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr7_+_141490017 | 0.03 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr5_+_64920543 | 0.03 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr4_-_175443943 | 0.03 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr16_+_82068585 | 0.03 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_+_9144626 | 0.03 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr13_+_23755054 | 0.03 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_+_19389814 | 0.03 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr9_+_123884038 | 0.03 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr3_+_63805017 | 0.03 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr14_-_92198403 | 0.03 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr1_+_235530675 | 0.03 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr3_-_196911002 | 0.03 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_-_159766528 | 0.03 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr10_-_22292613 | 0.03 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chrX_-_24045303 | 0.03 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr7_-_35013217 | 0.02 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr12_-_70093065 | 0.02 |
ENST00000553096.1
|
BEST3
|
bestrophin 3 |
| chr7_-_19748640 | 0.02 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr1_+_63073437 | 0.02 |
ENST00000593719.1
|
AL138847.1
|
CDNA FLJ26506 fis, clone KDN07105; Uncharacterized protein |
| chr4_-_85654615 | 0.02 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr6_+_143999185 | 0.02 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr12_-_11244912 | 0.02 |
ENST00000531678.1
|
TAS2R43
|
taste receptor, type 2, member 43 |
| chr2_+_46926048 | 0.02 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr11_-_85780853 | 0.02 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_+_7705398 | 0.02 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr3_-_193096600 | 0.02 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr2_+_161993412 | 0.02 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_68530697 | 0.02 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr12_-_70093190 | 0.02 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
| chr14_-_31926623 | 0.02 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr5_+_56471592 | 0.02 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr22_+_21128167 | 0.02 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chrX_+_135388147 | 0.02 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr17_+_57233087 | 0.02 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr8_-_7320974 | 0.02 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chrX_-_148571884 | 0.02 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr12_+_20963647 | 0.02 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_+_161123270 | 0.02 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_+_143999072 | 0.02 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr15_-_83474806 | 0.02 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr20_-_17539456 | 0.02 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr7_+_79765071 | 0.02 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chrM_+_14741 | 0.02 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr12_-_57328187 | 0.02 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr5_+_142286887 | 0.02 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_-_40657397 | 0.02 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_+_35856951 | 0.02 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr17_-_55911970 | 0.02 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr10_-_105845674 | 0.02 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr19_-_13068012 | 0.02 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr22_-_18111499 | 0.02 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr15_-_73925651 | 0.02 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr7_-_54826920 | 0.02 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr8_-_126963417 | 0.02 |
ENST00000500989.2
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr5_+_95997918 | 0.02 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr5_+_115177178 | 0.02 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr10_+_72194585 | 0.02 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr10_-_127464390 | 0.02 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chr9_-_97356075 | 0.02 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr2_+_33359473 | 0.02 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr8_-_101730061 | 0.02 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_151034734 | 0.02 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr9_+_108463234 | 0.02 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr7_-_18067478 | 0.02 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr3_+_238456 | 0.02 |
ENST00000427688.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr3_+_238427 | 0.02 |
ENST00000397491.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr14_+_75179840 | 0.02 |
ENST00000554590.1
ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1
|
FCF1 rRNA-processing protein |
| chr14_-_68159152 | 0.02 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr9_-_21305312 | 0.02 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr6_+_32938692 | 0.02 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr6_+_88299833 | 0.02 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr12_-_118796971 | 0.02 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_207095212 | 0.02 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
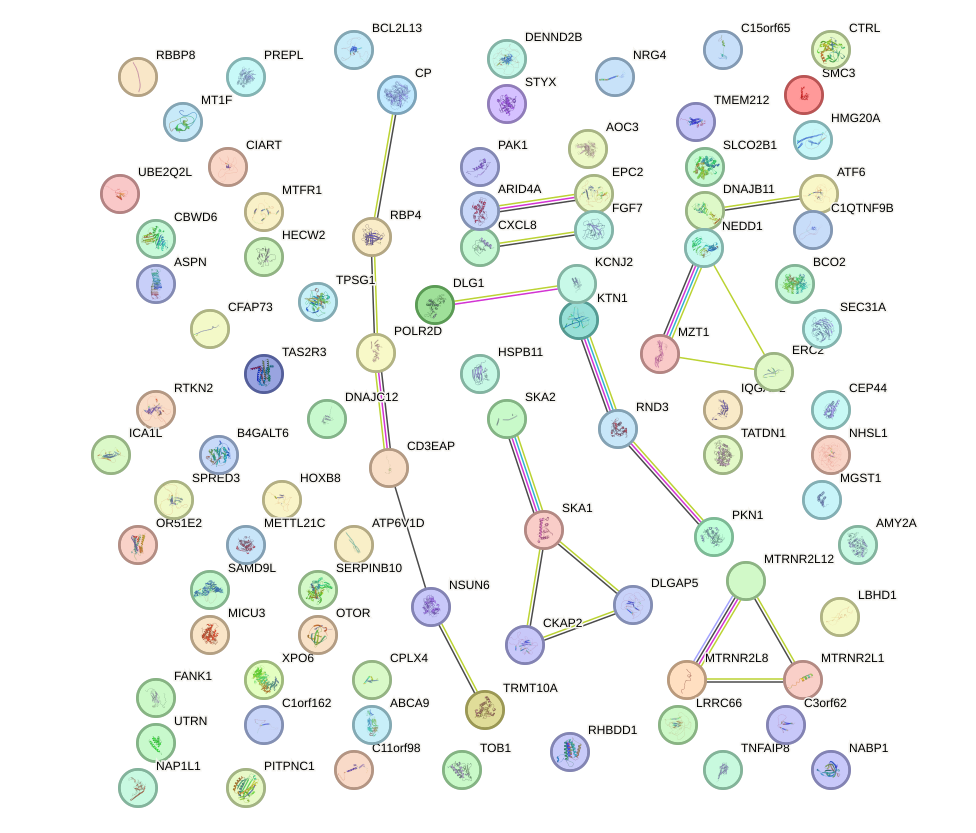
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |