Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for DLX3_EVX1_MEOX1
Z-value: 0.14
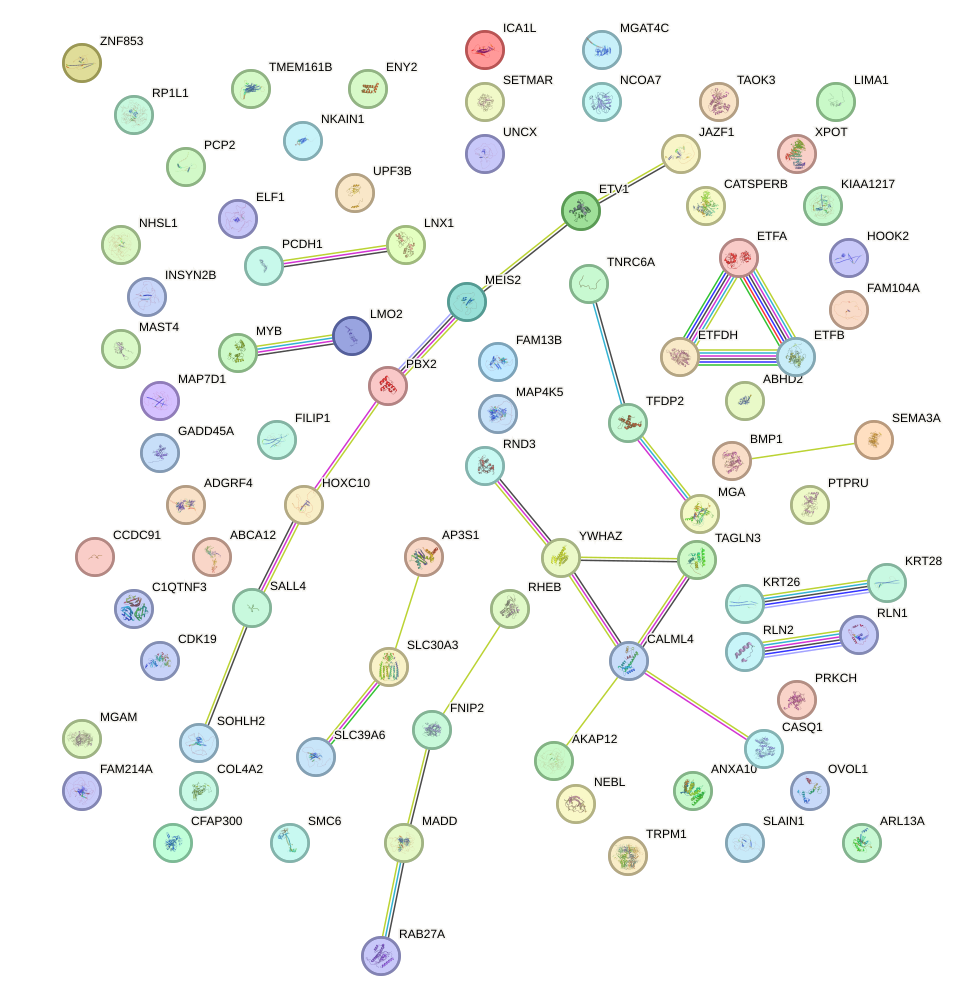
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | -0.88 | 1.2e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53133070 | 0.16 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_1922789 | 0.11 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr5_+_66300464 | 0.09 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_57233966 | 0.07 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr12_-_50643664 | 0.07 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr7_-_27169801 | 0.07 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr9_-_75488984 | 0.06 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr9_-_5304432 | 0.06 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr15_-_55562451 | 0.06 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr20_-_50418947 | 0.06 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_14029283 | 0.06 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr17_-_38928414 | 0.06 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr6_-_76203345 | 0.05 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr13_-_36788718 | 0.04 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr6_+_126221034 | 0.04 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_-_32157947 | 0.04 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr20_-_50418972 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_+_47293795 | 0.04 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr5_-_141249154 | 0.04 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr9_-_13165457 | 0.04 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr18_-_33709268 | 0.04 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr10_-_21186144 | 0.04 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr19_-_7698599 | 0.04 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr6_-_111136299 | 0.03 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr15_+_41913690 | 0.03 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr7_-_111424462 | 0.03 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr16_+_53412368 | 0.03 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr14_-_51027838 | 0.03 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_-_101963482 | 0.03 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_138866823 | 0.03 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr12_+_28410128 | 0.03 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chrX_+_100224676 | 0.03 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr14_+_61654271 | 0.03 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr7_-_111424506 | 0.03 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr6_+_151646800 | 0.03 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_+_32579271 | 0.03 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr3_+_4345287 | 0.02 |
ENST00000358950.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr13_+_110958124 | 0.02 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_-_141747950 | 0.02 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_+_101918153 | 0.02 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr16_+_24741013 | 0.02 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr7_+_6655225 | 0.02 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr6_-_76203454 | 0.02 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr18_-_68004529 | 0.02 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr4_+_77941685 | 0.02 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr12_+_54378849 | 0.02 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr16_+_14280742 | 0.02 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr7_+_1272522 | 0.02 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr4_+_169013666 | 0.02 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr15_-_55563072 | 0.02 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr6_+_135502501 | 0.02 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr20_-_50419055 | 0.02 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_+_64798826 | 0.02 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr2_-_17981462 | 0.02 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr13_+_78315466 | 0.02 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_+_32579321 | 0.02 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr12_-_94673956 | 0.02 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr17_-_38956205 | 0.02 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr4_+_159727272 | 0.02 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr1_+_202317815 | 0.02 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_83824449 | 0.02 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_-_151217166 | 0.02 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr1_+_68150744 | 0.02 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_-_68497657 | 0.02 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr7_-_28220354 | 0.02 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr2_-_151395525 | 0.02 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr7_-_33102399 | 0.02 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr6_+_47666275 | 0.02 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr3_+_111717511 | 0.02 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chrX_-_118986911 | 0.02 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr2_-_216003127 | 0.02 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr8_-_42358742 | 0.02 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr2_-_27498208 | 0.02 |
ENST00000424577.1
ENST00000426569.1 |
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr9_-_5339873 | 0.02 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr2_-_203735586 | 0.02 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr1_+_36621174 | 0.02 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr7_-_33102338 | 0.02 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr13_-_41593425 | 0.02 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr5_-_34043310 | 0.02 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr15_+_80351910 | 0.02 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr19_-_12889226 | 0.02 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr15_-_52971544 | 0.02 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr5_+_115177178 | 0.02 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_+_110346546 | 0.02 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr5_-_137374288 | 0.01 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr10_-_27529486 | 0.01 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr15_-_31393910 | 0.01 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_-_33913708 | 0.01 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr14_-_92247032 | 0.01 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr13_-_34250861 | 0.01 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr1_+_160160346 | 0.01 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr10_+_24738355 | 0.01 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr8_-_101963677 | 0.01 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_33686925 | 0.01 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_-_46187069 | 0.01 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr16_+_14280564 | 0.01 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr15_+_89631381 | 0.01 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr12_-_118628350 | 0.01 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr10_+_114710211 | 0.01 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_128231299 | 0.01 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr8_-_10512569 | 0.01 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr11_+_65554493 | 0.01 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr5_-_42811986 | 0.01 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_31666767 | 0.01 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr15_-_37393406 | 0.01 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr17_-_71223839 | 0.01 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr5_-_169407744 | 0.01 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr8_+_32579341 | 0.01 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr5_-_87516448 | 0.01 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr17_+_44679808 | 0.01 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr1_+_236958554 | 0.01 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr5_-_142780280 | 0.01 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_-_90085458 | 0.01 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chrX_+_43515467 | 0.01 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr9_+_130026756 | 0.01 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr9_+_116225999 | 0.01 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chrX_-_106243294 | 0.01 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr4_-_109684120 | 0.01 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr14_-_82000140 | 0.01 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr6_-_72130472 | 0.01 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr11_-_70672645 | 0.01 |
ENST00000423696.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr3_+_44840679 | 0.01 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr1_-_50889155 | 0.01 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr2_-_201753717 | 0.01 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr11_-_117748138 | 0.01 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr1_+_160160283 | 0.01 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr6_-_76072719 | 0.01 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_-_74486217 | 0.01 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_-_101690650 | 0.01 |
ENST00000543621.1
|
DNMBP
|
dynamin binding protein |
| chrX_-_106243451 | 0.01 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_+_111718036 | 0.01 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr8_-_103424916 | 0.01 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chrX_+_133930798 | 0.01 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr12_+_107712173 | 0.01 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr12_-_56693758 | 0.01 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr4_-_25865159 | 0.01 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_+_111415757 | 0.01 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr4_+_41614720 | 0.01 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_151034734 | 0.01 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chrX_+_123097014 | 0.01 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr1_+_225600404 | 0.01 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr15_+_93443419 | 0.01 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr4_-_69111401 | 0.01 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr3_+_111717600 | 0.01 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr1_-_101491319 | 0.01 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr6_+_142623758 | 0.01 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr10_+_118083919 | 0.01 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr2_-_55647057 | 0.01 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr6_-_111927062 | 0.01 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr8_-_103424986 | 0.01 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr3_-_39321512 | 0.01 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr13_+_78315348 | 0.01 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr17_-_39677971 | 0.01 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr7_-_99716940 | 0.01 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_+_146402346 | 0.01 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chrX_+_7137475 | 0.01 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr1_-_197115818 | 0.01 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr17_-_1418972 | 0.01 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr8_+_40010989 | 0.01 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr11_-_117747434 | 0.01 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chrM_+_12331 | 0.01 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_-_55646957 | 0.01 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr15_+_89631647 | 0.01 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr4_+_41614909 | 0.01 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_10532531 | 0.01 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chrM_+_7586 | 0.01 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr17_+_59489112 | 0.01 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr1_+_230193521 | 0.01 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr15_-_34875771 | 0.01 |
ENST00000267731.7
|
GOLGA8B
|
golgin A8 family, member B |
| chr1_+_62439037 | 0.01 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr11_+_107461804 | 0.01 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr7_+_99425633 | 0.01 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr6_-_111804905 | 0.01 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr8_-_103425047 | 0.01 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr15_+_58702742 | 0.01 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr6_-_33160231 | 0.01 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr2_-_182545603 | 0.01 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr16_-_66864806 | 0.01 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr8_-_49834299 | 0.01 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_-_14849470 | 0.01 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr3_-_123339343 | 0.01 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr4_-_36245561 | 0.01 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_-_174451370 | 0.01 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr8_-_49833978 | 0.01 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr4_+_174089904 | 0.01 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr14_+_100485712 | 0.01 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr15_+_63188009 | 0.01 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr15_+_80351977 | 0.01 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr12_-_719573 | 0.01 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr7_-_92777606 | 0.01 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr17_+_60536002 | 0.01 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr10_-_52008313 | 0.01 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr13_-_31191642 | 0.01 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr20_+_43029911 | 0.01 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr7_-_32338917 | 0.01 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr4_-_119759795 | 0.01 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr11_-_95657231 | 0.01 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr6_+_127898312 | 0.01 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr6_-_111136513 | 0.01 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |