Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
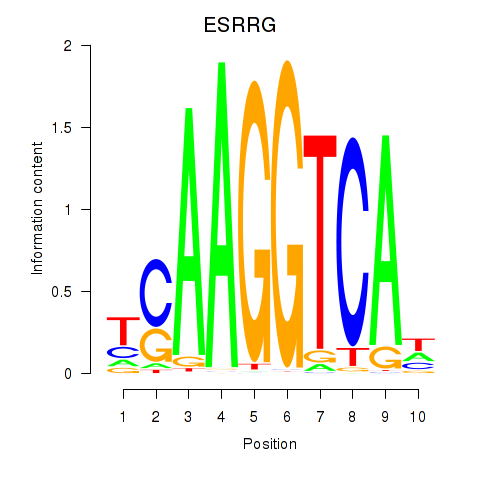
Results for ESRRB_ESRRG
Z-value: 0.84
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | estrogen related receptor beta |
|
ESRRG
|
ENSG00000196482.12 | estrogen related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRB | hg19_v2_chr14_+_76776957_76777061 | 0.89 | 1.1e-01 | Click! |
| ESRRG | hg19_v2_chr1_-_217311090_217311097 | -0.85 | 1.5e-01 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_109602039 | 0.92 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr11_+_66624527 | 0.76 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_-_790060 | 0.71 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr16_+_776936 | 0.69 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr11_-_63993601 | 0.64 |
ENST00000545812.1
ENST00000394547.3 ENST00000317459.6 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr14_-_103987679 | 0.57 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr11_-_118272610 | 0.56 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr11_+_67798114 | 0.55 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr16_+_777739 | 0.52 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr11_-_63993690 | 0.52 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr16_+_691792 | 0.51 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr7_-_150777920 | 0.47 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_-_1783633 | 0.42 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr18_-_56985776 | 0.41 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr11_-_47470682 | 0.39 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr10_-_76995675 | 0.39 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr10_-_76995769 | 0.38 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr16_+_777118 | 0.34 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr11_+_67798363 | 0.34 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr2_-_176046391 | 0.34 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr10_-_10504285 | 0.34 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr22_+_46546406 | 0.33 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr22_+_29702996 | 0.33 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr15_+_67813406 | 0.33 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr7_-_150777874 | 0.33 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_150777949 | 0.32 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr15_-_76352069 | 0.32 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr12_-_111358372 | 0.32 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr22_+_30115986 | 0.31 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr1_+_155179012 | 0.31 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr12_+_6644443 | 0.30 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_16083154 | 0.30 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_+_73455788 | 0.29 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr19_+_41699103 | 0.28 |
ENST00000597754.1
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr12_-_57039739 | 0.27 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr8_-_145752390 | 0.27 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr11_+_86667117 | 0.25 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr20_-_3748363 | 0.25 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr8_+_145149930 | 0.25 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr2_-_198364552 | 0.25 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr15_-_81616446 | 0.25 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr22_-_24110063 | 0.25 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr1_+_207262881 | 0.25 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_149889382 | 0.25 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr10_+_112257596 | 0.24 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr19_-_36019123 | 0.24 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_-_207119738 | 0.24 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr17_+_79679369 | 0.24 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr19_+_589893 | 0.23 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr1_-_235116495 | 0.23 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr5_+_175085033 | 0.23 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr11_+_66059339 | 0.23 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr17_-_1553346 | 0.22 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr12_+_121163538 | 0.22 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr22_+_30821732 | 0.21 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr1_+_36024107 | 0.21 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr11_+_22646739 | 0.21 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr14_-_102976135 | 0.21 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr19_-_15236562 | 0.21 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr6_+_89790490 | 0.21 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr15_+_67814008 | 0.21 |
ENST00000557807.1
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr22_+_50354104 | 0.20 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr16_+_28875126 | 0.20 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr20_-_3748416 | 0.20 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr17_-_79838860 | 0.20 |
ENST00000582866.1
|
RP11-498C9.15
|
RP11-498C9.15 |
| chr16_+_28874860 | 0.20 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr12_+_58176525 | 0.19 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr17_+_79679299 | 0.19 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_-_179322416 | 0.19 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr19_+_38307999 | 0.19 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr17_-_65235916 | 0.19 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr12_+_6643676 | 0.19 |
ENST00000396856.1
ENST00000396861.1 |
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr16_+_30937213 | 0.19 |
ENST00000427128.1
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr5_+_111964133 | 0.18 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chr6_-_26285737 | 0.18 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr16_-_29874211 | 0.18 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr19_+_2249308 | 0.18 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr6_-_33548006 | 0.18 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr1_+_16083098 | 0.18 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_16083123 | 0.18 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_-_3062465 | 0.18 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr22_-_30960876 | 0.18 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr16_+_777246 | 0.18 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr8_+_99956759 | 0.17 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr3_-_49823941 | 0.17 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr11_+_86502085 | 0.17 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr12_+_133067157 | 0.17 |
ENST00000261673.6
|
FBRSL1
|
fibrosin-like 1 |
| chr1_+_202995611 | 0.17 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr19_-_55574538 | 0.17 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr22_-_39239987 | 0.16 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr22_+_46546494 | 0.16 |
ENST00000396000.2
ENST00000262735.5 ENST00000420804.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr8_+_124429006 | 0.16 |
ENST00000522194.1
ENST00000523356.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr16_-_67190152 | 0.16 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr3_-_179322436 | 0.16 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr3_-_49726104 | 0.15 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_-_3859089 | 0.15 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr1_+_1260147 | 0.15 |
ENST00000343938.4
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr12_+_120875910 | 0.15 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr12_-_100656134 | 0.15 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr11_-_68611721 | 0.15 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr19_-_7293942 | 0.15 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr11_+_67798090 | 0.15 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr11_+_77899920 | 0.15 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr15_-_94614049 | 0.15 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr8_+_67405794 | 0.15 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr13_+_28519343 | 0.15 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr17_+_39868577 | 0.15 |
ENST00000329402.3
|
GAST
|
gastrin |
| chr17_-_73505961 | 0.15 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_3862206 | 0.15 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr12_+_51632638 | 0.14 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr6_-_43197189 | 0.14 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr22_+_40573921 | 0.14 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr11_-_3862059 | 0.14 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr19_-_15236470 | 0.14 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr8_+_99956662 | 0.14 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr2_+_217498105 | 0.14 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr19_+_45251804 | 0.14 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr1_-_211752073 | 0.14 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr16_-_67694597 | 0.14 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr17_+_900342 | 0.13 |
ENST00000327158.4
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast) |
| chr19_+_3185910 | 0.13 |
ENST00000588428.1
|
NCLN
|
nicalin |
| chr17_-_79792909 | 0.13 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr16_+_4674787 | 0.13 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr9_+_133978190 | 0.13 |
ENST00000372312.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr7_+_2443202 | 0.13 |
ENST00000258711.6
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr4_+_119512928 | 0.13 |
ENST00000567913.2
|
RP11-384K6.6
|
RP11-384K6.6 |
| chr3_-_156840776 | 0.13 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr16_-_89787360 | 0.13 |
ENST00000389386.3
|
VPS9D1
|
VPS9 domain containing 1 |
| chr4_-_1723040 | 0.13 |
ENST00000382936.3
ENST00000536901.1 ENST00000303277.2 |
TMEM129
|
transmembrane protein 129 |
| chr14_-_102976091 | 0.13 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr6_-_33547975 | 0.13 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr17_+_38975358 | 0.13 |
ENST00000436612.1
ENST00000301665.3 |
TMEM99
|
transmembrane protein 99 |
| chr17_+_79670386 | 0.13 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr6_-_97345689 | 0.13 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr11_+_62623544 | 0.13 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr3_-_10028366 | 0.12 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr11_+_62623621 | 0.12 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_+_1248547 | 0.12 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr18_-_56985873 | 0.12 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr18_-_43678241 | 0.12 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr12_-_53320245 | 0.12 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr5_-_140027175 | 0.12 |
ENST00000512088.1
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr11_+_65769946 | 0.12 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr16_+_31366455 | 0.12 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr6_+_30749649 | 0.12 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr12_+_6976687 | 0.12 |
ENST00000396705.5
|
TPI1
|
triosephosphate isomerase 1 |
| chr12_+_121163602 | 0.12 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr11_+_62623512 | 0.12 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_6411636 | 0.12 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr8_+_11188397 | 0.12 |
ENST00000382435.4
|
SLC35G5
|
solute carrier family 35, member G5 |
| chr19_-_3062881 | 0.12 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr16_-_69390209 | 0.12 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr15_-_77988485 | 0.11 |
ENST00000561030.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr22_+_30821784 | 0.11 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr17_+_74723031 | 0.11 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr9_-_140095186 | 0.11 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr16_-_57481278 | 0.11 |
ENST00000567751.1
ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1
|
cytokine induced apoptosis inhibitor 1 |
| chr17_+_74075263 | 0.11 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr2_+_219135115 | 0.11 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr19_-_3063099 | 0.11 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr1_+_33231268 | 0.11 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr16_-_19896832 | 0.11 |
ENST00000537135.1
ENST00000564449.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr11_+_63742050 | 0.11 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr8_+_66933779 | 0.11 |
ENST00000276570.5
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr15_+_43809797 | 0.11 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr19_+_18726786 | 0.11 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr7_+_76107444 | 0.11 |
ENST00000435861.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr19_+_46171464 | 0.11 |
ENST00000590918.1
ENST00000263281.3 ENST00000304207.8 |
GIPR
|
gastric inhibitory polypeptide receptor |
| chr11_+_122733011 | 0.11 |
ENST00000533709.1
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr4_-_21699380 | 0.11 |
ENST00000382148.3
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr19_+_29704142 | 0.11 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr3_-_49158218 | 0.11 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr7_-_105029812 | 0.11 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_-_153514241 | 0.11 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr22_+_30792980 | 0.11 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_-_20834586 | 0.10 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr3_-_52312337 | 0.10 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr3_-_52443799 | 0.10 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr13_-_111214015 | 0.10 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr8_+_101349823 | 0.10 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr19_-_49016418 | 0.10 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr20_+_3776371 | 0.10 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr11_-_64511575 | 0.10 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr21_-_46330545 | 0.10 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_-_96926313 | 0.10 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr11_-_6462210 | 0.10 |
ENST00000265983.3
|
HPX
|
hemopexin |
| chr9_+_131873227 | 0.10 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_+_42372764 | 0.10 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr16_-_70729496 | 0.10 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr19_-_40931891 | 0.10 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr6_-_13328050 | 0.10 |
ENST00000420456.1
|
TBC1D7
|
TBC1 domain family, member 7 |
| chr9_-_131872928 | 0.10 |
ENST00000455830.2
ENST00000393384.3 ENST00000318080.2 |
CRAT
|
carnitine O-acetyltransferase |
| chr20_+_1875942 | 0.10 |
ENST00000358771.4
|
SIRPA
|
signal-regulatory protein alpha |
| chr17_-_2169425 | 0.10 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr14_-_24911868 | 0.10 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr9_-_130889990 | 0.10 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr22_+_30792846 | 0.10 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_-_157108266 | 0.10 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr22_+_30163340 | 0.10 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr16_+_680932 | 0.09 |
ENST00000319070.2
|
WFIKKN1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 0.5 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.3 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.7 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.5 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.1 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.2 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.1 | 0.2 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.2 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:0002415 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0018194 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:1902869 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 1.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0052847 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |