Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
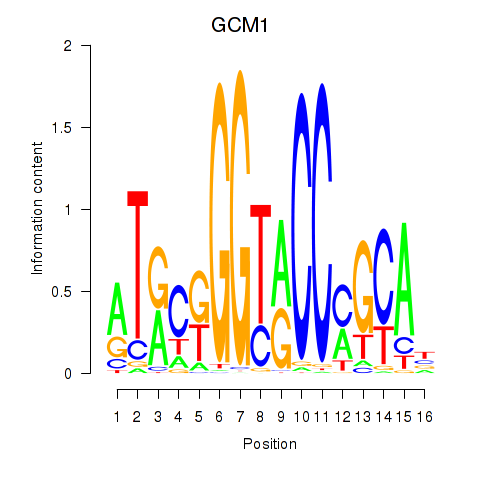
Results for GCM1
Z-value: 1.07
Transcription factors associated with GCM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM1
|
ENSG00000137270.10 | glial cells missing transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM1 | hg19_v2_chr6_-_53013620_53013644 | 0.54 | 4.6e-01 | Click! |
Activity profile of GCM1 motif
Sorted Z-values of GCM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_83776324 | 0.63 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr12_-_57824561 | 0.47 |
ENST00000448732.1
|
R3HDM2
|
R3H domain containing 2 |
| chr6_-_109761707 | 0.41 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr4_+_39640787 | 0.37 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr5_-_133702761 | 0.34 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr8_+_141522017 | 0.31 |
ENST00000518971.1
ENST00000519618.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr6_+_42928485 | 0.31 |
ENST00000372808.3
|
GNMT
|
glycine N-methyltransferase |
| chr11_+_13690249 | 0.30 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr1_-_40237020 | 0.29 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr12_+_96337061 | 0.28 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr15_-_71184724 | 0.28 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr8_+_141521386 | 0.28 |
ENST00000220913.5
ENST00000519533.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr10_-_101673782 | 0.27 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr3_+_32148106 | 0.27 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr2_+_149402009 | 0.26 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr4_+_56212505 | 0.26 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr2_+_208527094 | 0.26 |
ENST00000429730.1
|
AC079767.4
|
AC079767.4 |
| chr18_+_21529811 | 0.26 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr11_+_13690200 | 0.25 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr17_-_19619917 | 0.25 |
ENST00000325411.5
ENST00000350657.5 ENST00000433844.2 |
SLC47A2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2 |
| chr1_+_17634689 | 0.24 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr17_-_55911970 | 0.23 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr16_-_74734742 | 0.23 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr17_+_41005283 | 0.23 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr19_-_3480540 | 0.22 |
ENST00000215531.4
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr3_-_121468513 | 0.22 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr9_+_140125209 | 0.22 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr11_+_17756279 | 0.22 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr8_+_26434578 | 0.22 |
ENST00000493789.2
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr3_+_12329397 | 0.21 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr9_+_128510454 | 0.21 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr1_+_25071848 | 0.21 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr22_+_38864041 | 0.21 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr11_-_47470703 | 0.21 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr2_+_47596634 | 0.20 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chrX_-_84634737 | 0.20 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr12_-_39300071 | 0.20 |
ENST00000550863.1
|
CPNE8
|
copine VIII |
| chr4_+_76649797 | 0.20 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr3_+_32147997 | 0.20 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr12_-_6665200 | 0.20 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr1_+_24069952 | 0.19 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr14_-_105531759 | 0.19 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr16_+_67062996 | 0.19 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr3_+_130613226 | 0.19 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_+_120789223 | 0.19 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr20_+_60698180 | 0.19 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr9_+_17579084 | 0.19 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr18_+_21269556 | 0.19 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr3_+_130613001 | 0.18 |
ENST00000504948.1
ENST00000513801.1 ENST00000505072.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_+_92861653 | 0.18 |
ENST00000251739.5
ENST00000305866.5 ENST00000544910.1 ENST00000541136.1 ENST00000458530.1 ENST00000535481.1 ENST00000317751.6 |
CCDC132
|
coiled-coil domain containing 132 |
| chr3_-_121468602 | 0.18 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chrX_+_153672468 | 0.18 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr4_-_52904425 | 0.18 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr1_+_227751231 | 0.18 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr17_+_80517216 | 0.18 |
ENST00000531030.1
ENST00000526383.2 |
FOXK2
|
forkhead box K2 |
| chr12_-_56709674 | 0.18 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr4_-_119274121 | 0.17 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr15_-_66545995 | 0.17 |
ENST00000395614.1
ENST00000288745.3 ENST00000422354.1 ENST00000395625.2 ENST00000360698.4 ENST00000409699.2 |
MEGF11
|
multiple EGF-like-domains 11 |
| chrX_-_84634708 | 0.17 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr16_-_74734672 | 0.17 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr17_-_15902903 | 0.17 |
ENST00000486655.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr4_-_40859132 | 0.17 |
ENST00000543538.1
ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr7_+_64254766 | 0.17 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr1_+_161185032 | 0.17 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr8_-_144099795 | 0.17 |
ENST00000522060.1
ENST00000517833.1 ENST00000502167.2 ENST00000518831.1 |
RP11-273G15.2
|
RP11-273G15.2 |
| chr20_+_2854066 | 0.17 |
ENST00000455631.1
ENST00000216877.6 ENST00000399903.2 ENST00000358719.4 ENST00000431048.1 ENST00000425918.2 ENST00000430705.1 ENST00000318266.5 |
PTPRA
|
protein tyrosine phosphatase, receptor type, A |
| chr10_-_73975657 | 0.17 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr16_+_19729586 | 0.17 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr2_-_86564740 | 0.17 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr17_-_4269768 | 0.17 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr7_+_27779714 | 0.16 |
ENST00000265393.6
ENST00000409980.1 ENST00000433216.2 ENST00000396319.2 |
TAX1BP1
|
Tax1 (human T-cell leukemia virus type I) binding protein 1 |
| chr12_+_48225126 | 0.16 |
ENST00000550909.1
ENST00000550720.1 ENST00000548564.1 |
RP5-1057I20.2
|
RP5-1057I20.2 |
| chr7_-_37382683 | 0.16 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_25316707 | 0.16 |
ENST00000380665.3
|
CDCA2
|
cell division cycle associated 2 |
| chr18_-_18691739 | 0.16 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr2_+_233320827 | 0.15 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr10_-_134756030 | 0.15 |
ENST00000368586.5
ENST00000368582.2 |
TTC40
|
tetratricopeptide repeat domain 40 |
| chr6_-_41122063 | 0.15 |
ENST00000426005.2
ENST00000437044.2 ENST00000373127.4 |
TREML1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr19_-_37096139 | 0.15 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr1_-_160253998 | 0.15 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr11_+_35198118 | 0.15 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr10_-_75910539 | 0.15 |
ENST00000372745.1
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr16_+_67063142 | 0.15 |
ENST00000412916.2
|
CBFB
|
core-binding factor, beta subunit |
| chr6_+_52285131 | 0.15 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr3_-_107099454 | 0.15 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr7_+_129074266 | 0.14 |
ENST00000249344.2
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr1_+_1260598 | 0.14 |
ENST00000488011.1
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr7_+_94139105 | 0.14 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr19_-_12886327 | 0.14 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr16_+_333152 | 0.14 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr11_+_19138670 | 0.14 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr10_-_105677427 | 0.14 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr1_+_24069642 | 0.14 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr22_+_19705928 | 0.14 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr8_-_57232656 | 0.14 |
ENST00000396721.2
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr18_+_10526008 | 0.13 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr4_+_2627159 | 0.13 |
ENST00000382839.3
ENST00000324666.5 ENST00000545951.1 ENST00000502458.1 ENST00000505311.1 |
FAM193A
|
family with sequence similarity 193, member A |
| chr4_+_164415855 | 0.13 |
ENST00000508268.1
|
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr16_+_81478775 | 0.13 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr2_+_187350883 | 0.13 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr19_+_49977818 | 0.13 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr15_-_83735889 | 0.13 |
ENST00000379403.2
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr11_-_47470591 | 0.13 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr4_-_8073554 | 0.13 |
ENST00000510277.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr4_+_15683404 | 0.13 |
ENST00000422728.2
|
FAM200B
|
family with sequence similarity 200, member B |
| chr18_+_21269404 | 0.13 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr16_-_90096309 | 0.13 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr11_+_64058820 | 0.13 |
ENST00000422670.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr6_+_3068606 | 0.13 |
ENST00000259808.4
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr11_+_9406169 | 0.13 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr17_-_40913275 | 0.13 |
ENST00000589716.1
ENST00000360166.3 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr7_-_26904317 | 0.13 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr12_-_57824739 | 0.13 |
ENST00000347140.3
ENST00000402412.1 |
R3HDM2
|
R3H domain containing 2 |
| chr6_+_110012462 | 0.13 |
ENST00000441478.2
ENST00000230124.3 |
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
| chr10_-_31146615 | 0.13 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr5_-_142780280 | 0.12 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_-_113192065 | 0.12 |
ENST00000409750.1
|
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr1_-_222886526 | 0.12 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr10_-_105677886 | 0.12 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr17_+_41006095 | 0.12 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase, copper containing 3 |
| chr2_-_24583168 | 0.12 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr3_+_130612803 | 0.12 |
ENST00000510168.1
ENST00000508532.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_+_22676808 | 0.12 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_-_106810783 | 0.12 |
ENST00000283148.7
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr16_-_21170762 | 0.12 |
ENST00000261383.3
ENST00000415178.1 |
DNAH3
|
dynein, axonemal, heavy chain 3 |
| chr7_+_105172532 | 0.12 |
ENST00000257700.2
|
RINT1
|
RAD50 interactor 1 |
| chr8_+_104383728 | 0.12 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_-_8986474 | 0.12 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr14_+_50779029 | 0.12 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr5_-_127674883 | 0.12 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr7_+_23221438 | 0.12 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr22_-_42486747 | 0.12 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr2_-_106810742 | 0.12 |
ENST00000409501.3
ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr16_-_85722530 | 0.11 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr10_-_43762329 | 0.11 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr15_-_83736091 | 0.11 |
ENST00000261721.4
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr21_-_40555393 | 0.11 |
ENST00000380900.2
|
PSMG1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr16_+_3115611 | 0.11 |
ENST00000530890.1
ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32
|
interleukin 32 |
| chr4_-_99851766 | 0.11 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr15_+_78632666 | 0.11 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr2_+_73612858 | 0.11 |
ENST00000409009.1
ENST00000264448.6 ENST00000377715.1 |
ALMS1
|
Alstrom syndrome 1 |
| chr9_-_127952187 | 0.11 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr8_+_107670064 | 0.11 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr9_+_4985016 | 0.11 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr5_-_218251 | 0.11 |
ENST00000296824.3
|
CCDC127
|
coiled-coil domain containing 127 |
| chr2_-_197664366 | 0.11 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr5_-_137071756 | 0.11 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr20_-_45061695 | 0.11 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chr16_+_88872176 | 0.11 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr8_-_97247759 | 0.10 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr4_-_8073705 | 0.10 |
ENST00000514025.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr6_+_52285046 | 0.10 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr5_-_79287060 | 0.10 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr7_-_152373216 | 0.10 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr3_-_133380731 | 0.10 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr12_+_69864129 | 0.10 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chrX_-_63615297 | 0.10 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr12_-_69326940 | 0.10 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr12_+_32552451 | 0.10 |
ENST00000534526.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_-_24732368 | 0.10 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr14_+_39736299 | 0.10 |
ENST00000341502.5
ENST00000396158.2 ENST00000280083.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr6_-_56707943 | 0.10 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr11_+_118443098 | 0.10 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr5_+_122847781 | 0.10 |
ENST00000395412.1
ENST00000395411.1 ENST00000345990.4 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr17_+_15902694 | 0.10 |
ENST00000261647.5
ENST00000486880.2 |
TTC19
|
tetratricopeptide repeat domain 19 |
| chr11_+_73360024 | 0.09 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_-_110012380 | 0.09 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr4_+_15704679 | 0.09 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr4_-_5894777 | 0.09 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr14_+_77647966 | 0.09 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr2_-_70529126 | 0.09 |
ENST00000438759.1
ENST00000430566.1 |
FAM136A
|
family with sequence similarity 136, member A |
| chr3_+_12329358 | 0.09 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr16_+_67063262 | 0.09 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr9_+_130374537 | 0.09 |
ENST00000373302.3
ENST00000373299.1 |
STXBP1
|
syntaxin binding protein 1 |
| chr10_-_73976025 | 0.09 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr16_-_11485922 | 0.09 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr2_+_946543 | 0.09 |
ENST00000308624.5
ENST00000407292.1 |
SNTG2
|
syntrophin, gamma 2 |
| chr4_+_38511367 | 0.09 |
ENST00000507056.1
|
RP11-213G21.1
|
RP11-213G21.1 |
| chr11_+_111126707 | 0.09 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr17_+_34058639 | 0.09 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr12_+_42624050 | 0.09 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr6_-_74363636 | 0.09 |
ENST00000393019.3
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr9_-_99540328 | 0.09 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr11_+_31531291 | 0.09 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr5_+_122847908 | 0.09 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr14_+_39736582 | 0.08 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr1_-_36906474 | 0.08 |
ENST00000433045.2
|
OSCP1
|
organic solute carrier partner 1 |
| chr15_-_71407833 | 0.08 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr19_+_4769117 | 0.08 |
ENST00000540211.1
ENST00000317292.3 ENST00000586721.1 ENST00000592709.1 ENST00000588711.1 ENST00000589639.1 ENST00000591008.1 ENST00000592663.1 ENST00000588758.1 |
MIR7-3HG
|
MIR7-3 host gene (non-protein coding) |
| chr11_+_111412271 | 0.08 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr21_-_38445443 | 0.08 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_-_37382360 | 0.08 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_+_48611853 | 0.08 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr11_-_8190534 | 0.08 |
ENST00000309737.6
ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3
|
RIC3 acetylcholine receptor chaperone |
| chr1_+_27719148 | 0.08 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr12_+_11802753 | 0.08 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr5_-_892648 | 0.08 |
ENST00000483173.1
ENST00000435709.2 |
BRD9
|
bromodomain containing 9 |
| chr4_+_164415785 | 0.08 |
ENST00000513272.1
ENST00000513134.1 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr10_+_81466084 | 0.08 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr15_+_68117872 | 0.08 |
ENST00000380035.2
ENST00000389002.1 |
SKOR1
|
SKI family transcriptional corepressor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.2 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.3 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.2 | GO:0001812 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) positive regulation of type II hypersensitivity(GO:0002894) positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.0 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.6 | GO:0097484 | dendrite extension(GO:0097484) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 0.3 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.0 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |