Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GFI1B
Z-value: 0.41
Transcription factors associated with GFI1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1B
|
ENSG00000165702.8 | growth factor independent 1B transcriptional repressor |
Activity profile of GFI1B motif
Sorted Z-values of GFI1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_149688971 | 0.21 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr6_-_93433538 | 0.20 |
ENST00000404689.2
|
RP11-127B16.1
|
RP11-127B16.1 |
| chr19_-_16008880 | 0.16 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr14_-_80678512 | 0.15 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr2_-_208030295 | 0.14 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr21_-_43916433 | 0.14 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr21_-_43916296 | 0.14 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr14_-_23284675 | 0.14 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_95616933 | 0.13 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chrX_+_49832231 | 0.13 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr5_+_72509751 | 0.13 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr19_-_52307357 | 0.13 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr12_+_27619743 | 0.13 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr12_-_56756553 | 0.12 |
ENST00000398189.3
ENST00000541105.1 |
APOF
|
apolipoprotein F |
| chr19_+_44617511 | 0.12 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr8_-_102275131 | 0.12 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr2_+_233404429 | 0.12 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr17_+_46537697 | 0.11 |
ENST00000579972.1
|
RP11-433M22.2
|
RP11-433M22.2 |
| chr3_-_158450475 | 0.11 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr8_+_40018977 | 0.11 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr19_+_9203855 | 0.11 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr19_+_36602104 | 0.10 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr19_+_42037433 | 0.10 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr6_-_56707943 | 0.10 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr22_-_37880543 | 0.10 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_26225354 | 0.10 |
ENST00000360408.1
|
HIST1H3E
|
histone cluster 1, H3e |
| chr5_+_66254698 | 0.10 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_44040488 | 0.10 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr11_-_85430356 | 0.09 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr18_+_55712915 | 0.09 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_56814968 | 0.09 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr17_+_30089454 | 0.09 |
ENST00000577970.1
|
RP11-805L22.3
|
RP11-805L22.3 |
| chr6_+_131571535 | 0.08 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr12_-_70093235 | 0.08 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr7_+_120591170 | 0.08 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr4_-_909521 | 0.07 |
ENST00000511229.1
|
GAK
|
cyclin G associated kinase |
| chr12_+_96337061 | 0.07 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr7_+_134576317 | 0.07 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr6_+_71123107 | 0.07 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr9_-_100954910 | 0.07 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr8_+_11291429 | 0.07 |
ENST00000533578.1
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr22_+_19466980 | 0.07 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr17_-_79827808 | 0.07 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr6_+_116601330 | 0.07 |
ENST00000449314.1
ENST00000453463.1 |
RP1-93H18.1
|
RP1-93H18.1 |
| chr4_-_48116540 | 0.07 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr2_+_85646054 | 0.07 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr19_-_44123734 | 0.07 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr7_+_149535455 | 0.07 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr19_-_54604083 | 0.06 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr2_-_69180083 | 0.06 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr6_-_46620522 | 0.06 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr7_+_66093908 | 0.06 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr20_+_34679725 | 0.06 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr21_+_35014706 | 0.06 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr5_+_64920543 | 0.06 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr4_-_185694872 | 0.06 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr7_+_66800928 | 0.06 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr6_-_56708459 | 0.06 |
ENST00000370788.2
|
DST
|
dystonin |
| chr4_+_108852697 | 0.06 |
ENST00000508453.1
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr10_-_18948208 | 0.06 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr4_+_78829479 | 0.06 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr7_+_120590803 | 0.06 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr15_+_71228826 | 0.06 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_-_48908805 | 0.06 |
ENST00000273860.4
|
OCIAD2
|
OCIA domain containing 2 |
| chr2_+_152214098 | 0.06 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_201994042 | 0.06 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr7_+_79765071 | 0.06 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_-_161695042 | 0.06 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr8_-_28965684 | 0.06 |
ENST00000523130.1
|
KIF13B
|
kinesin family member 13B |
| chr1_-_49242553 | 0.06 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr9_-_104145795 | 0.06 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr12_+_100967420 | 0.06 |
ENST00000266754.5
ENST00000547754.1 |
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr12_-_70093111 | 0.06 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr15_+_48413211 | 0.06 |
ENST00000449382.2
|
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr11_-_33774944 | 0.06 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr6_+_12290586 | 0.06 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr21_+_35014783 | 0.06 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr19_+_57631411 | 0.05 |
ENST00000254181.4
ENST00000600940.1 |
USP29
|
ubiquitin specific peptidase 29 |
| chr2_+_242295658 | 0.05 |
ENST00000264042.3
ENST00000545004.1 ENST00000373287.4 |
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr15_-_72514866 | 0.05 |
ENST00000562997.1
|
PKM
|
pyruvate kinase, muscle |
| chr6_-_131384347 | 0.05 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_50474238 | 0.05 |
ENST00000399206.1
|
C14orf182
|
chromosome 14 open reading frame 182 |
| chr5_-_171404730 | 0.05 |
ENST00000518752.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr2_-_208030886 | 0.05 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_-_131384373 | 0.05 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chrX_+_7137475 | 0.05 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr4_-_156875003 | 0.05 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr6_+_90272488 | 0.05 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr6_-_131384412 | 0.05 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_-_129924468 | 0.05 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr18_+_56532053 | 0.05 |
ENST00000592452.1
|
ZNF532
|
zinc finger protein 532 |
| chr4_+_41362796 | 0.05 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr6_+_32937083 | 0.05 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr16_-_4838255 | 0.05 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr11_+_70116779 | 0.05 |
ENST00000253925.7
ENST00000389547.3 |
PPFIA1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr13_+_113623509 | 0.05 |
ENST00000535094.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr3_-_15540055 | 0.05 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr1_+_164529004 | 0.05 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_+_39663061 | 0.05 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr5_-_137071756 | 0.05 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr1_-_23751189 | 0.04 |
ENST00000374601.3
ENST00000450454.2 |
TCEA3
|
transcription elongation factor A (SII), 3 |
| chr10_-_129924611 | 0.04 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr7_-_139756791 | 0.04 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr20_+_32254286 | 0.04 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr1_-_197169672 | 0.04 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr1_-_45965525 | 0.04 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr12_-_13105081 | 0.04 |
ENST00000541128.1
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D |
| chr1_-_111970353 | 0.04 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr5_-_137475071 | 0.04 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr15_-_75918787 | 0.04 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
| chr10_+_94590910 | 0.04 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr10_-_13523073 | 0.04 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr12_-_28125638 | 0.04 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr6_+_71122974 | 0.04 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr10_+_114206956 | 0.04 |
ENST00000432306.1
ENST00000393077.2 |
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A |
| chr6_+_153019023 | 0.04 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
myc target 1 |
| chr17_+_47287749 | 0.04 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr2_-_176033066 | 0.04 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr21_-_22175341 | 0.04 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr15_+_48413169 | 0.04 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr16_+_4838412 | 0.04 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr15_+_67458861 | 0.04 |
ENST00000558428.1
ENST00000558827.1 |
SMAD3
|
SMAD family member 3 |
| chr17_+_75450099 | 0.04 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr12_-_112614506 | 0.04 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr12_+_117348742 | 0.04 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr5_+_109025067 | 0.04 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr2_+_54683419 | 0.04 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_49813090 | 0.04 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr20_+_16729003 | 0.04 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr5_-_138725560 | 0.04 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr3_-_112738565 | 0.04 |
ENST00000383675.2
ENST00000314400.5 |
C3orf17
|
chromosome 3 open reading frame 17 |
| chr3_-_112738490 | 0.04 |
ENST00000393857.2
|
C3orf17
|
chromosome 3 open reading frame 17 |
| chr2_+_9563769 | 0.04 |
ENST00000475482.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr10_+_24498060 | 0.04 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr18_+_11857439 | 0.04 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_+_35544968 | 0.03 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr4_+_119606523 | 0.03 |
ENST00000388822.5
ENST00000506780.1 ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr12_-_95397442 | 0.03 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr14_+_58765305 | 0.03 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr9_-_110540419 | 0.03 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr7_-_99332719 | 0.03 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr8_+_62200509 | 0.03 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr7_-_150721570 | 0.03 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr20_+_47835884 | 0.03 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr17_+_75449889 | 0.03 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr4_+_26585538 | 0.03 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr5_-_125930929 | 0.03 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr16_-_10912605 | 0.03 |
ENST00000299866.8
|
TVP23A
|
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chr13_-_27745936 | 0.03 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr1_-_220219775 | 0.03 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr3_-_33759699 | 0.03 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr12_+_50017327 | 0.03 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr8_-_10987745 | 0.03 |
ENST00000400102.3
|
AF131215.5
|
AF131215.5 |
| chr2_-_27938593 | 0.03 |
ENST00000379677.2
|
AC074091.13
|
Uncharacterized protein |
| chr13_+_111805980 | 0.03 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_-_182360498 | 0.03 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr2_-_201753717 | 0.03 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr6_-_146285455 | 0.03 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr16_+_50776021 | 0.03 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr7_-_100808843 | 0.03 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr1_-_182360918 | 0.03 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr3_-_47018219 | 0.03 |
ENST00000292314.2
ENST00000546280.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr5_+_53813536 | 0.03 |
ENST00000343017.6
ENST00000381410.4 ENST00000326277.3 |
SNX18
|
sorting nexin 18 |
| chr19_-_41934635 | 0.03 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr17_-_37764128 | 0.03 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr16_+_4838393 | 0.03 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr2_-_96874553 | 0.03 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr12_+_93861264 | 0.03 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr3_-_149688655 | 0.03 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr15_-_42500351 | 0.03 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr19_+_41770236 | 0.03 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr11_+_32112431 | 0.03 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr8_+_145159846 | 0.03 |
ENST00000532522.1
ENST00000527572.1 ENST00000527058.1 |
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chr1_-_9714644 | 0.03 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr4_+_26165074 | 0.03 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_21910775 | 0.03 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr1_+_23953817 | 0.03 |
ENST00000374555.3
|
MDS2
|
myelodysplastic syndrome 2 translocation associated |
| chr12_+_117176113 | 0.03 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr9_-_123239632 | 0.03 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr12_-_28124903 | 0.03 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr11_+_5617330 | 0.03 |
ENST00000278302.5
ENST00000424369.1 ENST00000507320.1 ENST00000380107.1 |
TRIM6
|
tripartite motif containing 6 |
| chr12_-_56221330 | 0.03 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr2_-_110371412 | 0.03 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr18_+_77905794 | 0.03 |
ENST00000587254.1
ENST00000586421.1 |
AC139100.2
|
Uncharacterized protein |
| chrX_+_16141667 | 0.03 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr4_+_56815102 | 0.03 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr19_+_33071974 | 0.03 |
ENST00000590247.2
ENST00000419343.3 ENST00000592786.1 ENST00000379316.3 |
PDCD5
|
programmed cell death 5 |
| chr11_+_5617952 | 0.03 |
ENST00000354852.5
|
TRIM6-TRIM34
|
TRIM6-TRIM34 readthrough |
| chr5_+_79703823 | 0.03 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr15_+_45544426 | 0.03 |
ENST00000347644.3
ENST00000560438.1 |
SLC28A2
|
solute carrier family 28 (concentrative nucleoside transporter), member 2 |
| chr15_-_70994612 | 0.03 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr13_+_76362974 | 0.03 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr6_+_90272027 | 0.03 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr16_+_15039742 | 0.03 |
ENST00000537062.1
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr17_+_75450075 | 0.03 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr7_-_100808394 | 0.03 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr7_+_134576151 | 0.03 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr1_+_39796810 | 0.03 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr3_-_93781750 | 0.03 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr11_+_93861993 | 0.03 |
ENST00000227638.3
ENST00000436171.2 |
PANX1
|
pannexin 1 |
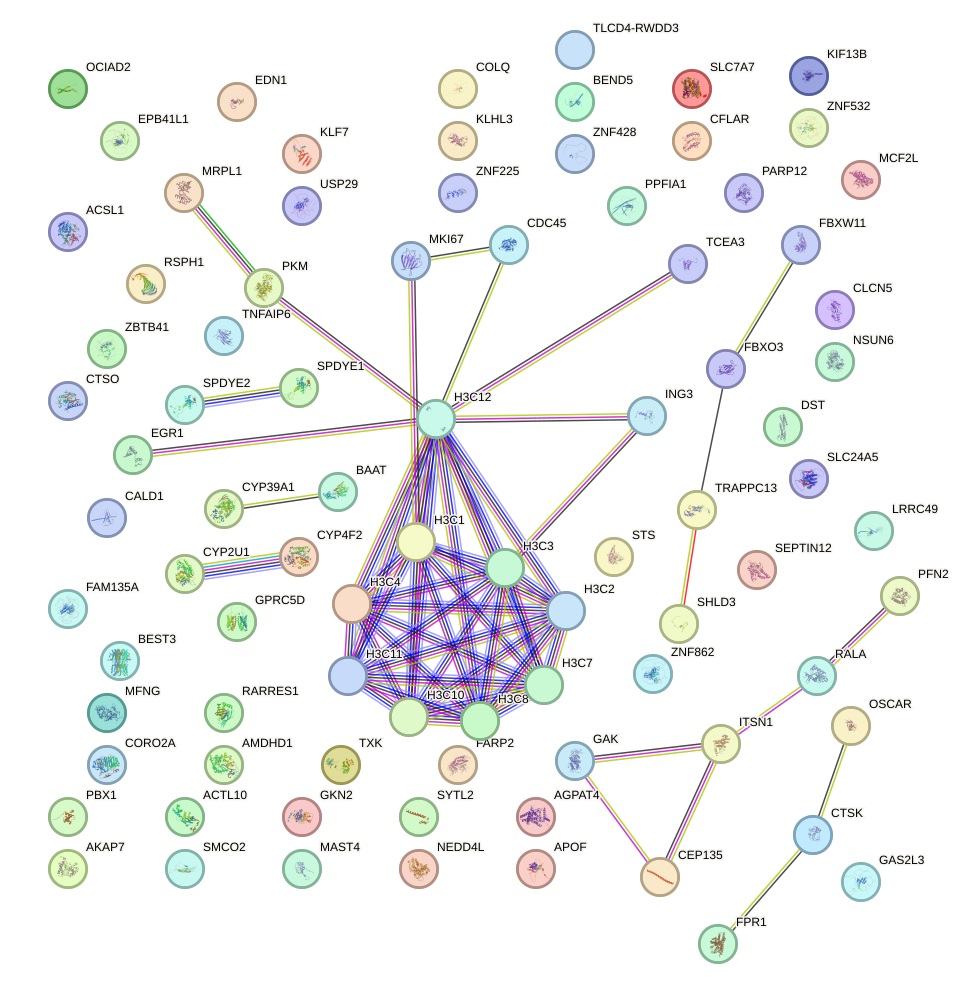
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0060585 | nitric oxide transport(GO:0030185) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:2000359 | negative regulation of fertilization(GO:0060467) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |