Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GUGCAAA
Z-value: 0.77
miRNA associated with seed GUGCAAA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-19a-3p
|
MIMAT0000073 |
|
hsa-miR-19b-3p
|
MIMAT0000074 |
Activity profile of GUGCAAA motif
Sorted Z-values of GUGCAAA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_68165657 | 0.49 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr14_-_34931458 | 0.29 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr6_-_132272504 | 0.26 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr4_+_75230853 | 0.24 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chrX_-_135056216 | 0.23 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr10_+_65281123 | 0.22 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr4_-_100867864 | 0.22 |
ENST00000442697.2
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chrX_-_131352152 | 0.22 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr2_+_64681219 | 0.21 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr18_-_51751132 | 0.21 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chrX_-_108976521 | 0.20 |
ENST00000469796.2
ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr8_-_67525473 | 0.20 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr8_-_103668114 | 0.20 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr7_+_23636992 | 0.19 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr17_-_27278304 | 0.19 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr2_+_189156389 | 0.19 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr11_-_59436453 | 0.18 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr18_-_5296001 | 0.18 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr13_-_107187462 | 0.18 |
ENST00000245323.4
|
EFNB2
|
ephrin-B2 |
| chr2_+_70142189 | 0.18 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr1_+_26438289 | 0.18 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr6_+_138188551 | 0.18 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_+_27793197 | 0.18 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr5_+_95997918 | 0.18 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr1_+_112162381 | 0.18 |
ENST00000433097.1
ENST00000369709.3 ENST00000436150.2 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr20_-_52210368 | 0.18 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr3_-_156877997 | 0.17 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr10_+_180987 | 0.17 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chrX_-_34675391 | 0.17 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr1_-_244615425 | 0.17 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chr10_+_95256356 | 0.16 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr10_-_118764862 | 0.16 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr20_+_47662805 | 0.16 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr15_+_52311398 | 0.16 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr1_-_243418344 | 0.16 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_+_193091080 | 0.16 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr12_-_122907091 | 0.16 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr11_-_19263145 | 0.15 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr11_-_95522907 | 0.15 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr12_+_64845660 | 0.15 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr13_-_27745936 | 0.15 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr12_-_93323013 | 0.15 |
ENST00000322349.8
|
EEA1
|
early endosome antigen 1 |
| chr12_+_69004619 | 0.15 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr17_-_62658186 | 0.15 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr5_-_90679145 | 0.15 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr17_+_30264014 | 0.15 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr2_-_202316260 | 0.15 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr1_-_93426998 | 0.14 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr10_+_86088381 | 0.14 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr15_-_73925651 | 0.14 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr15_+_63481668 | 0.14 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr7_+_35840542 | 0.14 |
ENST00000435235.1
ENST00000399034.2 ENST00000350320.6 ENST00000469679.2 |
SEPT7
|
septin 7 |
| chr15_-_52861394 | 0.14 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr1_-_91487013 | 0.14 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chrX_+_131157290 | 0.14 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr2_-_153032484 | 0.13 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr4_+_85504075 | 0.13 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr5_-_78809950 | 0.13 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_+_197687071 | 0.13 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr4_+_57774042 | 0.13 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr8_+_87354945 | 0.13 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_114562172 | 0.13 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr19_+_35521572 | 0.13 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr1_-_51984908 | 0.13 |
ENST00000371730.2
|
EPS15
|
epidermal growth factor receptor pathway substrate 15 |
| chr2_+_46926048 | 0.13 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr4_+_55524085 | 0.13 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr2_+_172950227 | 0.13 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr17_+_57784826 | 0.12 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr6_-_108279369 | 0.12 |
ENST00000369002.4
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chrY_+_15016725 | 0.12 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr20_-_62462566 | 0.12 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr18_-_61089665 | 0.12 |
ENST00000238497.5
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr4_+_140374961 | 0.12 |
ENST00000305626.5
|
RAB33B
|
RAB33B, member RAS oncogene family |
| chr1_-_70671216 | 0.12 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr11_+_69924397 | 0.12 |
ENST00000355303.5
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr6_-_134639180 | 0.12 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_49338748 | 0.12 |
ENST00000559471.1
|
SECISBP2L
|
SECIS binding protein 2-like |
| chr5_-_150460539 | 0.12 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr7_-_28220354 | 0.12 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr5_-_40798263 | 0.12 |
ENST00000296800.4
ENST00000397128.2 |
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr15_+_72947079 | 0.11 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr14_+_31028329 | 0.11 |
ENST00000206595.6
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr11_-_10315741 | 0.11 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr14_-_57735528 | 0.11 |
ENST00000340918.7
ENST00000413566.2 |
EXOC5
|
exocyst complex component 5 |
| chr2_+_231577532 | 0.11 |
ENST00000258418.5
|
CAB39
|
calcium binding protein 39 |
| chr3_-_9291063 | 0.11 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr3_+_67048721 | 0.11 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr16_-_47007545 | 0.11 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chrX_-_19905703 | 0.11 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr5_-_133968529 | 0.11 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr13_+_97874574 | 0.11 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr4_+_139936905 | 0.11 |
ENST00000280614.2
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr8_+_59323823 | 0.11 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr7_-_95064264 | 0.11 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr9_-_123964114 | 0.10 |
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family |
| chr12_+_72148614 | 0.10 |
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family |
| chr10_+_111767720 | 0.10 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr17_+_65821780 | 0.10 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr18_+_20513782 | 0.10 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr14_+_96829814 | 0.10 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr1_-_222885770 | 0.10 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr15_+_40532058 | 0.10 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr16_-_46723066 | 0.10 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chrX_+_72783026 | 0.10 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr4_-_5894777 | 0.10 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr6_+_16129308 | 0.10 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr11_+_33278811 | 0.10 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr14_-_53619816 | 0.10 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr2_+_103236004 | 0.10 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chrX_+_21392873 | 0.10 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr12_+_79258547 | 0.10 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr15_+_78730531 | 0.10 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr2_-_242212227 | 0.10 |
ENST00000427007.1
ENST00000458564.1 ENST00000452065.1 ENST00000427183.2 ENST00000426343.1 ENST00000422080.1 ENST00000449504.1 ENST00000449864.1 ENST00000391975.1 |
HDLBP
|
high density lipoprotein binding protein |
| chr19_-_31840438 | 0.10 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr14_-_45431091 | 0.10 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr11_+_18344106 | 0.10 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr2_+_264869 | 0.10 |
ENST00000272067.6
ENST00000272065.5 ENST00000407983.3 |
ACP1
|
acid phosphatase 1, soluble |
| chr15_-_100258029 | 0.10 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr3_+_171758344 | 0.09 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr1_+_24069642 | 0.09 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr3_+_57261743 | 0.09 |
ENST00000288266.3
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chrX_+_117480036 | 0.09 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chrX_+_16804544 | 0.09 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr2_-_39664405 | 0.09 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr17_-_5372271 | 0.09 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr10_-_27529716 | 0.09 |
ENST00000375897.3
ENST00000396271.3 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_-_169104651 | 0.09 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr3_-_182698381 | 0.09 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr14_-_81687197 | 0.09 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr1_+_211433275 | 0.09 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr7_+_91875508 | 0.09 |
ENST00000265742.3
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
| chr2_-_69614373 | 0.09 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr18_+_9913977 | 0.09 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr11_-_74109422 | 0.09 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr5_-_94620239 | 0.09 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_+_23728198 | 0.09 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr3_+_180630090 | 0.09 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr2_+_173292301 | 0.09 |
ENST00000264106.6
ENST00000375221.2 ENST00000343713.4 |
ITGA6
|
integrin, alpha 6 |
| chrX_+_41192595 | 0.09 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr1_+_57110972 | 0.09 |
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr4_-_10118469 | 0.09 |
ENST00000499869.2
|
WDR1
|
WD repeat domain 1 |
| chr9_+_19408999 | 0.09 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr11_+_34073195 | 0.09 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr5_-_1882858 | 0.08 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr15_+_28623784 | 0.08 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr8_-_12612962 | 0.08 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr2_+_9346892 | 0.08 |
ENST00000281419.3
ENST00000315273.4 |
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr22_+_40390930 | 0.08 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr1_-_169337176 | 0.08 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr20_+_42086525 | 0.08 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr7_-_27205136 | 0.08 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr2_-_201828356 | 0.08 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr1_+_229406847 | 0.08 |
ENST00000366690.4
|
RAB4A
|
RAB4A, member RAS oncogene family |
| chr4_+_128703295 | 0.08 |
ENST00000296464.4
ENST00000508549.1 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr7_-_45960850 | 0.08 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr1_-_71546690 | 0.08 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr8_-_17104356 | 0.08 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr3_+_178866199 | 0.08 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chrX_-_3631635 | 0.08 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr11_+_120894781 | 0.08 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr12_-_102513843 | 0.08 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr10_-_104178857 | 0.08 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_-_185286461 | 0.08 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr10_-_32636106 | 0.08 |
ENST00000263062.8
ENST00000319778.6 |
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr13_+_80055284 | 0.08 |
ENST00000218652.7
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr7_-_79082867 | 0.08 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_-_16870704 | 0.08 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr3_+_150804676 | 0.08 |
ENST00000474524.1
ENST00000273432.4 |
MED12L
|
mediator complex subunit 12-like |
| chr1_+_101361626 | 0.08 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_-_132073210 | 0.08 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr7_-_75368248 | 0.07 |
ENST00000434438.2
ENST00000336926.6 |
HIP1
|
huntingtin interacting protein 1 |
| chr5_+_98104978 | 0.07 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chrX_+_146993449 | 0.07 |
ENST00000218200.8
ENST00000370471.3 ENST00000370477.1 |
FMR1
|
fragile X mental retardation 1 |
| chr14_+_90863327 | 0.07 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr17_-_80606304 | 0.07 |
ENST00000392325.4
|
WDR45B
|
WD repeat domain 45B |
| chr8_+_67976593 | 0.07 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr4_-_85418103 | 0.07 |
ENST00000515820.2
|
NKX6-1
|
NK6 homeobox 1 |
| chr12_-_22697343 | 0.07 |
ENST00000446597.1
ENST00000536386.1 ENST00000396028.2 ENST00000545552.1 ENST00000544930.1 ENST00000333957.4 |
C2CD5
|
C2 calcium-dependent domain containing 5 |
| chr1_+_28099683 | 0.07 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr10_+_92980517 | 0.07 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr6_-_109703663 | 0.07 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr2_-_183903133 | 0.07 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr13_+_49550015 | 0.07 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr20_-_524455 | 0.07 |
ENST00000349736.5
ENST00000217244.3 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr3_+_152017181 | 0.07 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr19_+_41768401 | 0.07 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr13_-_50367057 | 0.07 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr7_-_111846435 | 0.07 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_+_64751433 | 0.07 |
ENST00000238856.4
ENST00000422803.1 ENST00000238855.7 |
AFTPH
|
aftiphilin |
| chr2_-_65357225 | 0.07 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr20_-_36793774 | 0.07 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chr6_+_121756809 | 0.07 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr5_+_86564739 | 0.07 |
ENST00000456692.2
ENST00000512763.1 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr5_-_156486120 | 0.07 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr1_-_202113805 | 0.07 |
ENST00000272217.2
|
ARL8A
|
ADP-ribosylation factor-like 8A |
| chr5_+_56205081 | 0.07 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr1_-_46598284 | 0.07 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr16_-_77468945 | 0.07 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr14_-_50698276 | 0.07 |
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila) |
| chr5_+_172483347 | 0.07 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
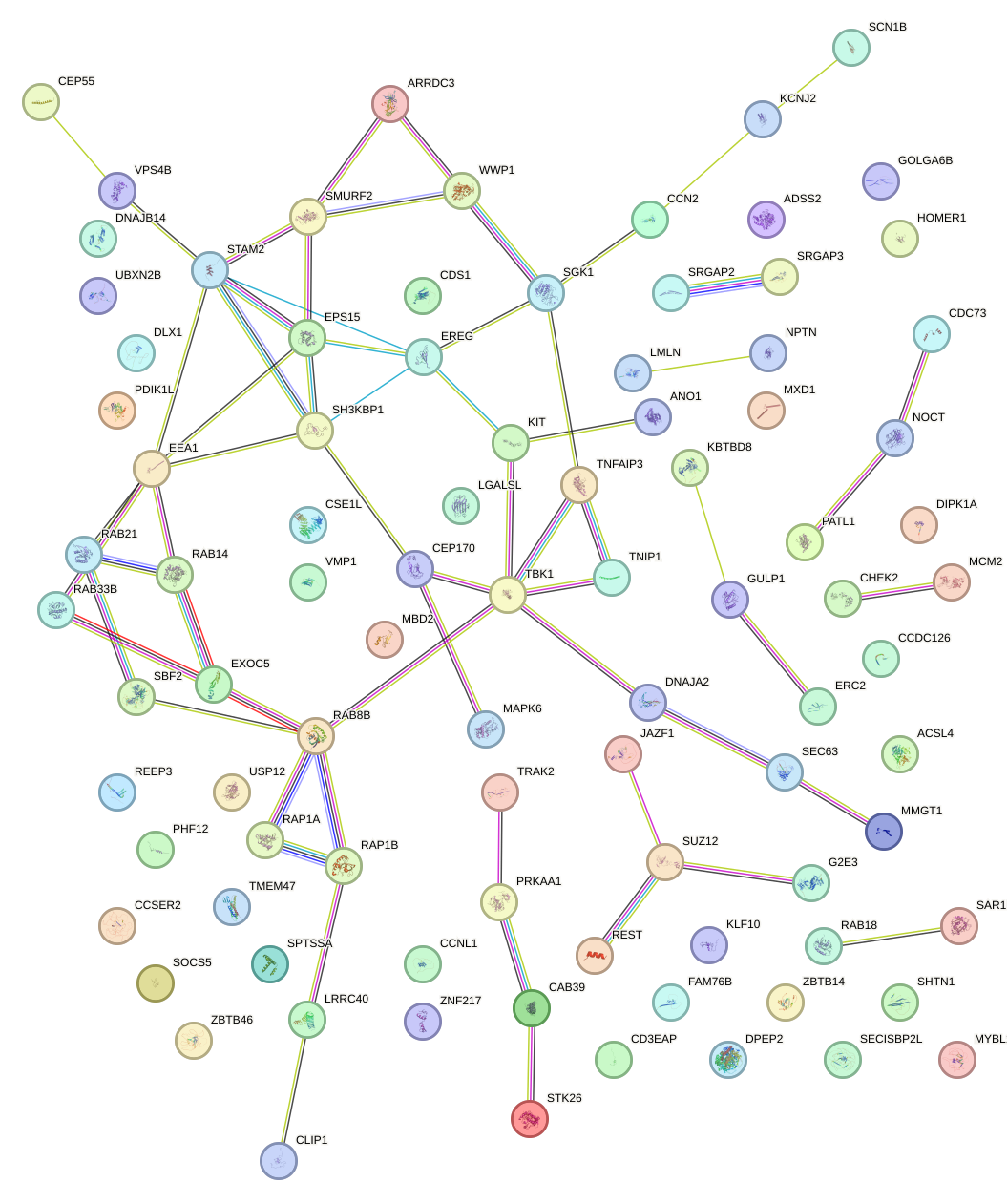
Network of associatons between targets according to the STRING database.
First level regulatory network of GUGCAAA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.2 | GO:2000349 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1902869 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:1903181 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.0 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.2 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 0.2 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.1 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |