Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HAND1
Z-value: 0.22
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.2 | heart and neural crest derivatives expressed 1 |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_54405773 | 0.10 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr19_+_507299 | 0.09 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr21_-_16125773 | 0.07 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr19_+_18492973 | 0.07 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr17_-_64216748 | 0.07 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr20_-_1165117 | 0.07 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr16_-_28621353 | 0.07 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr16_-_28608364 | 0.06 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr19_+_36359341 | 0.06 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chrX_+_49832231 | 0.05 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr19_-_10213335 | 0.05 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr14_+_96722152 | 0.05 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr22_+_25003606 | 0.05 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr22_+_25003568 | 0.04 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr16_-_28621298 | 0.04 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_-_63330842 | 0.04 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr17_+_45908974 | 0.04 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr14_+_96722539 | 0.04 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr4_-_40632757 | 0.03 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr17_+_6347761 | 0.03 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr2_+_98262497 | 0.03 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr20_-_1165319 | 0.03 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr12_-_7261772 | 0.03 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr2_+_210443993 | 0.03 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_-_45908875 | 0.03 |
ENST00000351111.2
ENST00000414011.1 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr3_-_43147549 | 0.03 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chrX_+_15767971 | 0.03 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr11_-_62477103 | 0.03 |
ENST00000532818.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr2_-_25475120 | 0.03 |
ENST00000380746.4
ENST00000402667.1 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr7_+_141490017 | 0.03 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr2_-_159237472 | 0.03 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr20_+_34020827 | 0.03 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr12_-_62653903 | 0.03 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr3_+_141103634 | 0.03 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_+_70364667 | 0.03 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr22_+_25003626 | 0.02 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_+_66624280 | 0.02 |
ENST00000585484.1
|
RP11-118B18.1
|
RP11-118B18.1 |
| chr17_+_6347729 | 0.02 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr14_+_24422795 | 0.02 |
ENST00000313250.5
ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr22_+_24999114 | 0.02 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_+_207262170 | 0.02 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr22_-_39928823 | 0.02 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr2_+_13677795 | 0.02 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr12_+_68042495 | 0.02 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr19_-_36304201 | 0.02 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr16_-_28621312 | 0.02 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_-_62476694 | 0.02 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_39942322 | 0.02 |
ENST00000449889.1
ENST00000465293.1 |
JUP
|
junction plakoglobin |
| chr17_-_43025005 | 0.02 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr1_+_215747118 | 0.02 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr4_+_22999152 | 0.02 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr1_+_28261621 | 0.02 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr7_-_140714739 | 0.02 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr1_-_214638146 | 0.02 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr15_+_74509530 | 0.02 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr2_-_228582709 | 0.02 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr12_+_68042517 | 0.02 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr22_-_24093267 | 0.02 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr15_-_76304731 | 0.02 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr12_+_110338063 | 0.01 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr7_+_76139833 | 0.01 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr6_+_35996859 | 0.01 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr19_+_47421933 | 0.01 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr20_+_816695 | 0.01 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr2_+_210444298 | 0.01 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr16_+_90089008 | 0.01 |
ENST00000268699.4
|
GAS8
|
growth arrest-specific 8 |
| chr1_+_110036699 | 0.01 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr1_+_155911480 | 0.01 |
ENST00000368318.3
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4 |
| chr19_-_37701386 | 0.01 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr11_+_369804 | 0.01 |
ENST00000329962.6
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr5_+_143584814 | 0.01 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr4_-_40631859 | 0.01 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr17_+_78193443 | 0.01 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr4_-_40632140 | 0.01 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_-_61781750 | 0.01 |
ENST00000582026.1
|
STRADA
|
STE20-related kinase adaptor alpha |
| chr6_+_6588316 | 0.01 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr12_+_53848549 | 0.01 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr22_+_31518938 | 0.01 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr19_-_13044494 | 0.01 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr3_-_43147431 | 0.01 |
ENST00000441964.1
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr11_-_72432950 | 0.01 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_-_9811595 | 0.01 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr4_+_41614720 | 0.01 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_-_140714430 | 0.01 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr17_+_7836398 | 0.01 |
ENST00000565740.1
|
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr9_+_35805527 | 0.01 |
ENST00000421267.1
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr2_+_210444142 | 0.01 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr17_-_34890037 | 0.01 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr2_-_27718052 | 0.01 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr17_+_53342311 | 0.01 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr10_+_99344071 | 0.01 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr11_+_118826999 | 0.01 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr6_-_112194484 | 0.01 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr3_-_20227720 | 0.01 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr12_+_54379569 | 0.01 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr17_-_9725388 | 0.00 |
ENST00000399363.4
|
RP11-477N12.3
|
Putative germ cell-specific gene 1-like protein 2 |
| chr7_+_96634850 | 0.00 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr7_-_81399355 | 0.00 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_-_208031943 | 0.00 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_-_25162188 | 0.00 |
ENST00000302922.3
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr6_-_39282221 | 0.00 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr12_+_67663056 | 0.00 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr12_+_49717019 | 0.00 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr4_-_40632605 | 0.00 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr19_+_45165545 | 0.00 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr2_-_42588289 | 0.00 |
ENST00000468711.1
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr9_-_123555655 | 0.00 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr16_-_89556942 | 0.00 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr10_+_103113802 | 0.00 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr4_+_76481258 | 0.00 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr10_-_104211294 | 0.00 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr16_+_70328680 | 0.00 |
ENST00000563206.1
ENST00000451014.3 ENST00000568625.1 |
DDX19B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B |
| chr10_+_99344104 | 0.00 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr3_+_148415571 | 0.00 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
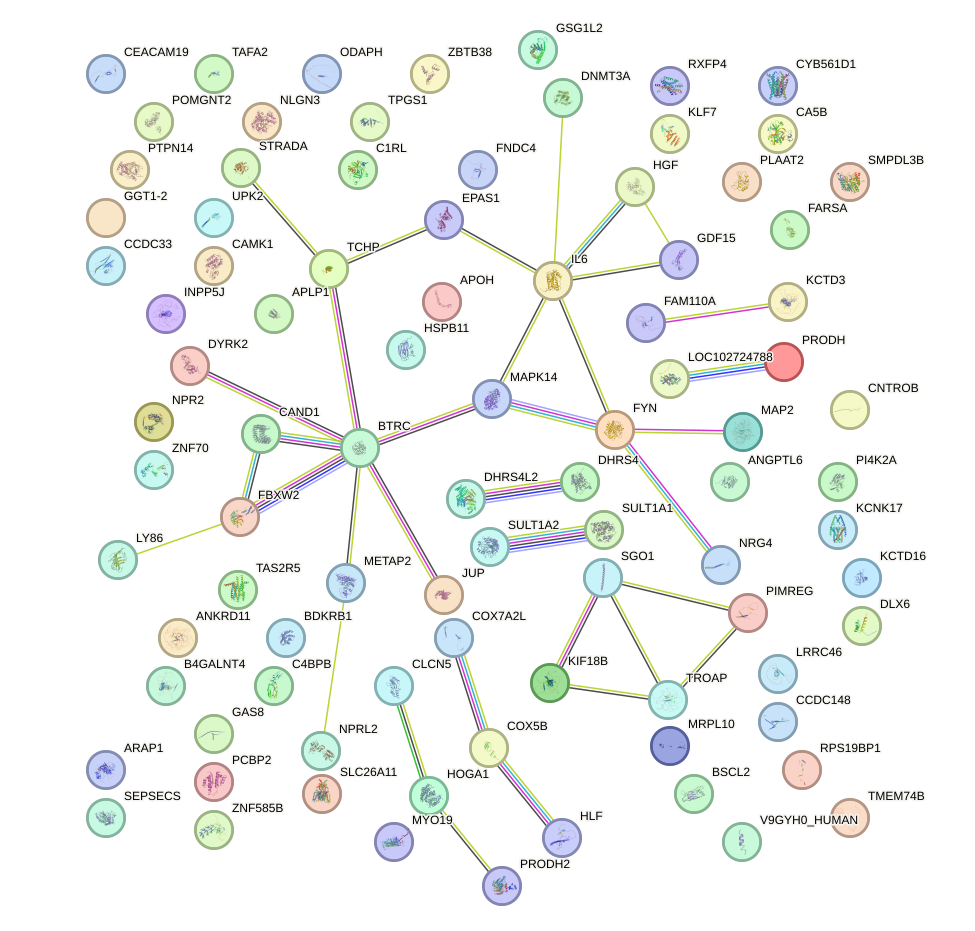
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |