Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HESX1
Z-value: 0.43
Transcription factors associated with HESX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HESX1
|
ENSG00000163666.4 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HESX1 | hg19_v2_chr3_-_57260377_57260549 | 0.44 | 5.6e-01 | Click! |
Activity profile of HESX1 motif
Sorted Z-values of HESX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26273144 | 0.33 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr2_+_90198535 | 0.16 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr2_+_90192768 | 0.14 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr1_-_43638168 | 0.12 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr12_-_49259643 | 0.11 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr12_-_49582593 | 0.11 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_-_46703069 | 0.11 |
ENST00000538237.1
ENST00000274793.7 |
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr15_+_80364901 | 0.10 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr8_+_97274119 | 0.10 |
ENST00000455950.2
|
PTDSS1
|
phosphatidylserine synthase 1 |
| chr6_+_43739697 | 0.09 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr9_+_95362539 | 0.09 |
ENST00000375579.3
|
CENPP
|
centromere protein P |
| chr4_-_89619386 | 0.09 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr20_-_62493217 | 0.08 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr2_+_176957619 | 0.08 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr10_+_90672113 | 0.08 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr8_+_71581565 | 0.08 |
ENST00000408926.3
ENST00000520030.1 |
XKR9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr6_-_31138439 | 0.08 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr2_+_90458201 | 0.08 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr8_-_97273807 | 0.07 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr19_+_34919257 | 0.07 |
ENST00000246548.4
ENST00000590048.2 |
UBA2
|
ubiquitin-like modifier activating enzyme 2 |
| chr4_+_15683404 | 0.07 |
ENST00000422728.2
|
FAM200B
|
family with sequence similarity 200, member B |
| chr21_-_38445443 | 0.07 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr20_-_40247002 | 0.07 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr1_-_161337662 | 0.06 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr22_+_20105259 | 0.06 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr19_+_37407212 | 0.06 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr22_+_44351419 | 0.06 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr1_-_40237020 | 0.06 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr20_+_35807512 | 0.06 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr17_+_47572647 | 0.06 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr10_+_103348031 | 0.06 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr11_-_66206260 | 0.06 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr12_+_10331605 | 0.06 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr3_+_9944303 | 0.06 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr12_-_121476750 | 0.05 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr14_-_24664540 | 0.05 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr1_+_47799446 | 0.05 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr5_+_52285144 | 0.05 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr2_+_3705785 | 0.05 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr2_+_230787201 | 0.05 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr2_-_197036289 | 0.05 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr20_+_35807449 | 0.05 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr10_-_128110441 | 0.05 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr1_-_54637997 | 0.05 |
ENST00000536061.1
|
AL357673.1
|
CDNA: FLJ21031 fis, clone CAE07336; HCG1780521; Uncharacterized protein |
| chr5_-_122759032 | 0.05 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr17_+_27573875 | 0.05 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr16_-_30134441 | 0.05 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr16_+_84733575 | 0.05 |
ENST00000219473.7
ENST00000563892.1 ENST00000562283.1 ENST00000570191.1 ENST00000569038.1 ENST00000570053.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr4_+_144303093 | 0.05 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_-_15168624 | 0.05 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr3_+_130648842 | 0.05 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_45553296 | 0.05 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr1_-_109968973 | 0.05 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr5_-_179780312 | 0.05 |
ENST00000253778.8
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr2_-_128051670 | 0.04 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr21_-_34852304 | 0.04 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr3_-_131753830 | 0.04 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr2_-_61765315 | 0.04 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr12_-_123717643 | 0.04 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr13_-_108870623 | 0.04 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr6_+_109416684 | 0.04 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr4_+_175204865 | 0.04 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr1_-_1356628 | 0.04 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr20_+_35807714 | 0.04 |
ENST00000373632.4
|
RPN2
|
ribophorin II |
| chr13_-_52378231 | 0.04 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr22_+_44351301 | 0.04 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr20_-_5485166 | 0.04 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr22_+_18111594 | 0.04 |
ENST00000399782.1
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr8_-_101734170 | 0.04 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr14_-_22938665 | 0.04 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr22_+_26565440 | 0.04 |
ENST00000404234.3
ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L
|
seizure related 6 homolog (mouse)-like |
| chr16_+_57220193 | 0.04 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr17_-_57184064 | 0.03 |
ENST00000262294.7
|
TRIM37
|
tripartite motif containing 37 |
| chr1_+_165600083 | 0.03 |
ENST00000367889.3
|
MGST3
|
microsomal glutathione S-transferase 3 |
| chr12_+_65218352 | 0.03 |
ENST00000539867.1
ENST00000544457.1 ENST00000539120.1 |
TBC1D30
|
TBC1 domain family, member 30 |
| chr21_-_38445011 | 0.03 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr15_+_22892663 | 0.03 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chrX_+_118602363 | 0.03 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr2_+_220071490 | 0.03 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr6_+_56820018 | 0.03 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr5_-_159546396 | 0.03 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr1_-_26372602 | 0.03 |
ENST00000374278.3
ENST00000374276.3 |
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr16_+_57279248 | 0.03 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr20_+_1115821 | 0.03 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_-_169337176 | 0.03 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr20_+_18269121 | 0.03 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr7_-_95025661 | 0.03 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr13_+_108870714 | 0.03 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr18_-_54318353 | 0.03 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr5_-_88180342 | 0.03 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr8_-_101963482 | 0.03 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr7_+_23637118 | 0.03 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr8_+_124084899 | 0.03 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr6_+_52285131 | 0.03 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr1_+_110009150 | 0.03 |
ENST00000401021.3
|
SYPL2
|
synaptophysin-like 2 |
| chr1_-_31661000 | 0.03 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr1_+_156338993 | 0.03 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr22_+_19419425 | 0.03 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr20_-_60982330 | 0.03 |
ENST00000279101.5
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr4_+_6784401 | 0.03 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr4_-_144826682 | 0.03 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr8_-_97173020 | 0.03 |
ENST00000287020.5
|
GDF6
|
growth differentiation factor 6 |
| chr11_+_118443098 | 0.03 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr2_-_160654745 | 0.03 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr1_-_205290865 | 0.03 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr9_+_1051481 | 0.03 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr2_+_186603355 | 0.03 |
ENST00000343098.5
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr6_+_56819773 | 0.03 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr20_-_40247133 | 0.03 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr13_+_100741269 | 0.03 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr15_+_80351910 | 0.03 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr8_-_101734308 | 0.03 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr16_+_46918235 | 0.03 |
ENST00000340124.4
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr10_-_75634326 | 0.03 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_-_202645612 | 0.02 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr7_-_75988321 | 0.02 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr6_+_52285046 | 0.02 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr12_+_131438443 | 0.02 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chrX_-_7895479 | 0.02 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr12_-_752786 | 0.02 |
ENST00000433832.2
ENST00000542920.1 |
NINJ2
|
ninjurin 2 |
| chr16_-_57219926 | 0.02 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr1_+_15256230 | 0.02 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr2_-_43823119 | 0.02 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr4_+_156588350 | 0.02 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr10_+_121652204 | 0.02 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr4_-_48782259 | 0.02 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr4_+_160203650 | 0.02 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr19_-_36004543 | 0.02 |
ENST00000339686.3
ENST00000447113.2 ENST00000440396.1 |
DMKN
|
dermokine |
| chr9_+_21440440 | 0.02 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr6_+_56819895 | 0.02 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr6_-_155635583 | 0.02 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr1_+_169337172 | 0.02 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr14_-_90798273 | 0.02 |
ENST00000357904.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chrX_+_105969893 | 0.02 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr16_+_19421803 | 0.02 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr1_-_165738072 | 0.02 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr2_+_96931834 | 0.02 |
ENST00000488633.1
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1 |
| chr18_-_44702668 | 0.02 |
ENST00000256433.3
|
IER3IP1
|
immediate early response 3 interacting protein 1 |
| chr12_-_121019165 | 0.02 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr2_+_233320827 | 0.02 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr2_-_43823093 | 0.02 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr3_-_42846021 | 0.02 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr21_+_38445539 | 0.02 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr17_+_55163075 | 0.02 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr19_+_47634039 | 0.02 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr11_-_4414880 | 0.02 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr6_+_116937636 | 0.02 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr12_+_123717967 | 0.02 |
ENST00000536130.1
ENST00000546132.1 |
C12orf65
|
chromosome 12 open reading frame 65 |
| chr11_+_111126707 | 0.02 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr1_-_165738085 | 0.02 |
ENST00000464650.1
ENST00000392129.6 |
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr2_+_234216454 | 0.02 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr4_+_129349188 | 0.02 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr17_-_57184260 | 0.02 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr6_-_83775489 | 0.02 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr7_+_115850547 | 0.02 |
ENST00000358204.4
ENST00000455989.1 ENST00000537767.1 |
TES
|
testis derived transcript (3 LIM domains) |
| chr9_+_37120536 | 0.02 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr1_+_46016703 | 0.01 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr1_-_203320617 | 0.01 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr12_+_13197218 | 0.01 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr1_-_20446020 | 0.01 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr8_+_79428539 | 0.01 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr12_-_112546547 | 0.01 |
ENST00000547133.1
|
NAA25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr21_-_38445470 | 0.01 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_+_83956312 | 0.01 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr11_+_64323098 | 0.01 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chrX_+_135730373 | 0.01 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chrX_-_131623043 | 0.01 |
ENST00000421707.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr7_-_37024665 | 0.01 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr7_-_99679324 | 0.01 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr22_+_31892373 | 0.01 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr3_-_131736593 | 0.01 |
ENST00000514999.1
|
CPNE4
|
copine IV |
| chr7_+_72742178 | 0.01 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr16_-_57219966 | 0.01 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr7_+_12727250 | 0.01 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr17_+_19282064 | 0.01 |
ENST00000603493.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_-_7389942 | 0.01 |
ENST00000296742.7
ENST00000502583.1 |
CAGE1
|
cancer antigen 1 |
| chr17_-_32906388 | 0.01 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr19_+_41698927 | 0.01 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chrX_+_135730297 | 0.01 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr17_-_31404 | 0.01 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr21_+_33784670 | 0.01 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr15_-_31453359 | 0.01 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_+_127228399 | 0.01 |
ENST00000000233.5
ENST00000415666.1 |
ARF5
|
ADP-ribosylation factor 5 |
| chr2_-_128399706 | 0.01 |
ENST00000426981.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr4_+_83956237 | 0.01 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr3_-_9885626 | 0.01 |
ENST00000424438.1
ENST00000433555.1 ENST00000427174.1 ENST00000418713.1 ENST00000433535.2 ENST00000383820.5 ENST00000433972.1 |
RPUSD3
|
RNA pseudouridylate synthase domain containing 3 |
| chr3_+_46921732 | 0.01 |
ENST00000418619.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr7_+_99746514 | 0.01 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr9_+_37079888 | 0.01 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr6_+_116421976 | 0.01 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr5_+_40679584 | 0.01 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_-_51058005 | 0.01 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr1_+_151372010 | 0.01 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr8_+_109455845 | 0.01 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr2_-_61765732 | 0.01 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr16_-_14724057 | 0.01 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr8_+_146277764 | 0.01 |
ENST00000331434.6
|
C8orf33
|
chromosome 8 open reading frame 33 |
| chr1_+_3614591 | 0.01 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr3_-_46735132 | 0.01 |
ENST00000415953.1
|
ALS2CL
|
ALS2 C-terminal like |
| chr5_-_41071392 | 0.01 |
ENST00000399564.4
|
MROH2B
|
maestro heat-like repeat family member 2B |
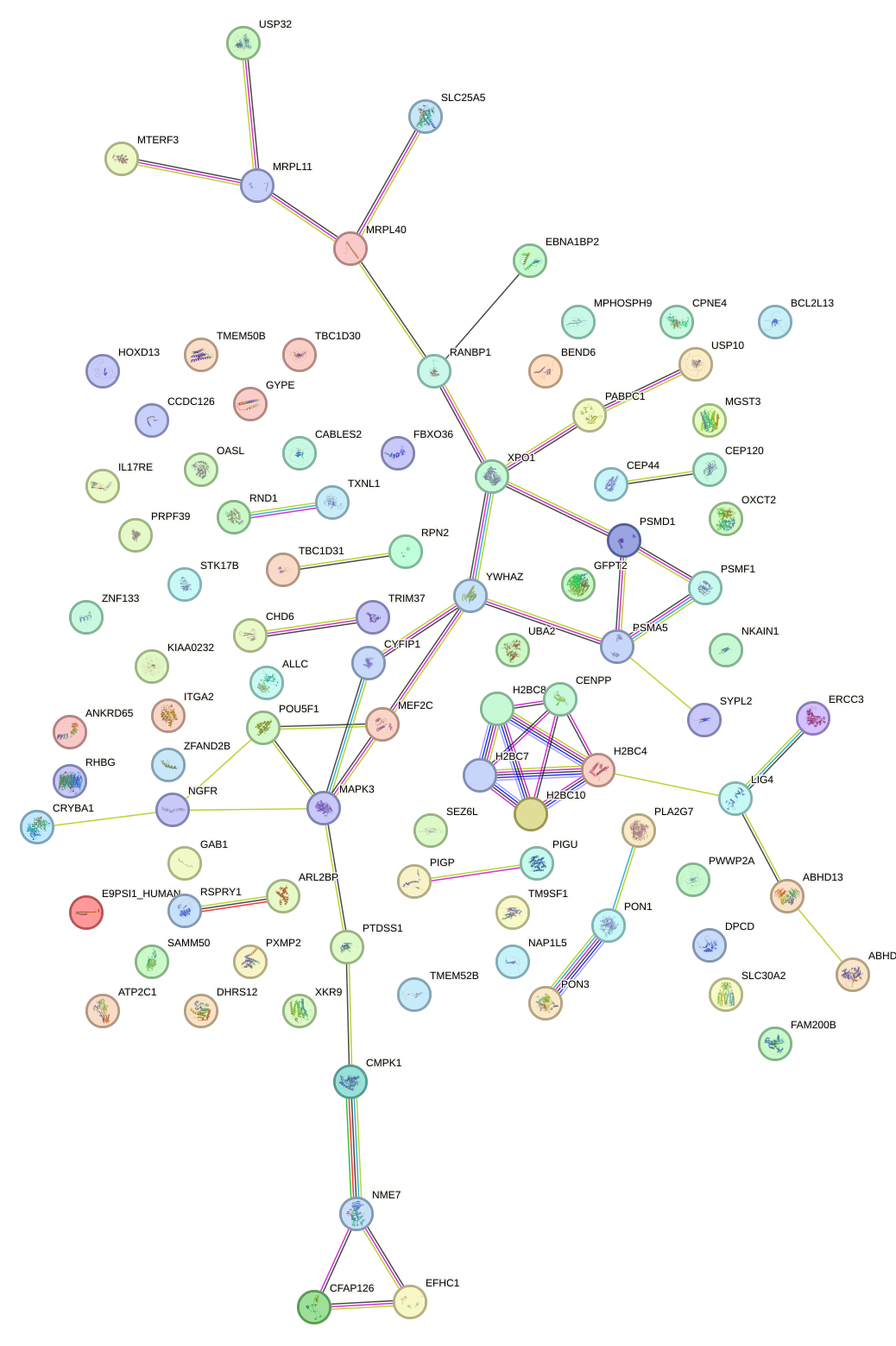
Network of associatons between targets according to the STRING database.
First level regulatory network of HESX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |