Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HMX1
Z-value: 0.26
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg19_v2_chr4_-_8873531_8873543 | 0.24 | 7.6e-01 | Click! |
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_99258625 | 0.13 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr3_-_107941209 | 0.13 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr2_+_130939235 | 0.10 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr17_-_79212884 | 0.10 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr11_+_66624527 | 0.09 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_+_41305612 | 0.09 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_41305330 | 0.08 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_6464502 | 0.08 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr12_-_49365501 | 0.08 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr8_-_103876340 | 0.07 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr16_-_776431 | 0.07 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr17_+_79679369 | 0.07 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr10_-_99531709 | 0.07 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr19_+_6464243 | 0.07 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr5_-_172662197 | 0.07 |
ENST00000521848.1
|
NKX2-5
|
NK2 homeobox 5 |
| chr17_+_79679299 | 0.07 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr14_+_52164675 | 0.06 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr12_-_54778444 | 0.06 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr17_-_41132088 | 0.06 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr6_-_31550192 | 0.06 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr5_-_172662303 | 0.06 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr14_+_102027688 | 0.06 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr7_-_559853 | 0.06 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr17_+_38465441 | 0.06 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr17_+_79213039 | 0.06 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr16_-_67190152 | 0.06 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr12_-_54778244 | 0.05 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr11_-_417308 | 0.05 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr5_-_172662230 | 0.05 |
ENST00000424406.2
|
NKX2-5
|
NK2 homeobox 5 |
| chr1_-_41328018 | 0.05 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr14_+_56584414 | 0.05 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr2_-_130939115 | 0.05 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chr5_+_141016508 | 0.05 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr22_-_39268192 | 0.05 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr8_+_145438870 | 0.05 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr17_-_15244894 | 0.05 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr19_-_1401486 | 0.05 |
ENST00000252288.2
ENST00000447102.3 |
GAMT
|
guanidinoacetate N-methyltransferase |
| chr17_-_73874654 | 0.05 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr19_-_10687948 | 0.05 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_417388 | 0.05 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr9_-_131940526 | 0.05 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr5_-_180230830 | 0.05 |
ENST00000427865.2
ENST00000514283.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_-_54778471 | 0.05 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_+_41305627 | 0.05 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr15_-_41166414 | 0.05 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr17_+_7284365 | 0.05 |
ENST00000311668.2
|
TNK1
|
tyrosine kinase, non-receptor, 1 |
| chrX_+_47444613 | 0.04 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr19_-_45579762 | 0.04 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr21_+_46875424 | 0.04 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr19_+_41305740 | 0.04 |
ENST00000596517.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr9_+_137967268 | 0.04 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr21_+_46875395 | 0.04 |
ENST00000355480.5
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr20_+_57464200 | 0.04 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr17_+_79990058 | 0.04 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_-_6052463 | 0.04 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr11_+_64692143 | 0.04 |
ENST00000164133.2
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr17_-_78450398 | 0.04 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr6_+_41606176 | 0.04 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr16_+_83986827 | 0.04 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr9_-_100000957 | 0.04 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr14_+_23564700 | 0.04 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr1_-_207095212 | 0.04 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chrX_-_153285251 | 0.04 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr3_-_128840604 | 0.03 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr3_+_127391769 | 0.03 |
ENST00000393363.3
ENST00000232744.8 ENST00000453791.2 |
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr12_-_49259643 | 0.03 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr3_-_49066811 | 0.03 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr22_-_39268308 | 0.03 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr6_+_44095263 | 0.03 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr17_+_74722912 | 0.03 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr1_-_153949751 | 0.03 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chrX_-_100914781 | 0.03 |
ENST00000431597.1
ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2
|
armadillo repeat containing, X-linked 2 |
| chrX_-_153285395 | 0.03 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_+_77751931 | 0.03 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chr5_-_58335281 | 0.03 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_+_826136 | 0.03 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr10_-_99258135 | 0.03 |
ENST00000327238.10
ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr16_+_67226019 | 0.03 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr19_-_7939319 | 0.03 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr9_+_140083099 | 0.03 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr17_-_1619535 | 0.03 |
ENST00000573075.1
ENST00000574306.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr22_-_32022280 | 0.03 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr17_-_79876010 | 0.03 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr1_+_95583479 | 0.03 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chrX_+_22050546 | 0.03 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_-_177713788 | 0.03 |
ENST00000280193.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr19_-_10687983 | 0.03 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_+_74723031 | 0.03 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr19_-_4124079 | 0.02 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr17_-_36831156 | 0.02 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr15_+_40763150 | 0.02 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr22_-_29196546 | 0.02 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr17_+_42836329 | 0.02 |
ENST00000200557.6
|
ADAM11
|
ADAM metallopeptidase domain 11 |
| chr8_-_38854332 | 0.02 |
ENST00000520152.1
ENST00000522142.1 |
TM2D2
|
TM2 domain containing 2 |
| chr1_-_32110467 | 0.02 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr15_+_89346699 | 0.02 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chrX_-_153744507 | 0.02 |
ENST00000442929.1
ENST00000426266.1 ENST00000359889.5 ENST00000369641.3 ENST00000447601.2 ENST00000434658.2 |
FAM3A
|
family with sequence similarity 3, member A |
| chr9_-_130331297 | 0.02 |
ENST00000373312.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr12_-_7077125 | 0.02 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr1_+_38512799 | 0.02 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr1_+_160336851 | 0.02 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr17_-_1619491 | 0.02 |
ENST00000570416.1
ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr19_-_42721819 | 0.02 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr8_+_103876528 | 0.02 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr1_-_17338386 | 0.02 |
ENST00000341676.5
ENST00000452699.1 |
ATP13A2
|
ATPase type 13A2 |
| chr11_+_60699222 | 0.02 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chr20_+_49348081 | 0.02 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr12_-_15374328 | 0.02 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr12_-_42631529 | 0.02 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr1_-_201476274 | 0.02 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr1_-_17338267 | 0.02 |
ENST00000326735.8
|
ATP13A2
|
ATPase type 13A2 |
| chr8_+_145691411 | 0.02 |
ENST00000301332.2
|
KIFC2
|
kinesin family member C2 |
| chr9_-_136024721 | 0.02 |
ENST00000393160.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chrX_+_37545012 | 0.02 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr1_+_9648921 | 0.02 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr12_+_57482665 | 0.02 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr11_-_76155700 | 0.02 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr18_+_34124507 | 0.02 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr16_+_29472707 | 0.02 |
ENST00000565290.1
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr11_-_67275542 | 0.02 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr22_+_37447771 | 0.02 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr12_+_1099675 | 0.02 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr5_+_139027877 | 0.02 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr12_+_53440753 | 0.02 |
ENST00000379902.3
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_-_49318715 | 0.02 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr11_+_45944190 | 0.02 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr7_+_12726623 | 0.02 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chrX_+_53123314 | 0.02 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chr4_-_80993854 | 0.02 |
ENST00000346652.6
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr9_-_127952032 | 0.02 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr3_-_18466787 | 0.02 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr17_-_62097927 | 0.02 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr9_-_23821273 | 0.02 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_+_55987998 | 0.02 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr2_+_234602305 | 0.02 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr7_-_156803329 | 0.02 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr16_+_3062457 | 0.02 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr9_-_140082983 | 0.02 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr22_+_50781723 | 0.02 |
ENST00000359139.3
ENST00000395741.3 ENST00000395744.3 |
PPP6R2
|
protein phosphatase 6, regulatory subunit 2 |
| chr1_-_154946825 | 0.02 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr16_+_30210552 | 0.01 |
ENST00000338971.5
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr2_+_30454390 | 0.01 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr2_-_220436248 | 0.01 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr16_-_58034357 | 0.01 |
ENST00000562909.1
|
ZNF319
|
zinc finger protein 319 |
| chr11_-_65308082 | 0.01 |
ENST00000532661.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr15_-_38852251 | 0.01 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr6_-_33679452 | 0.01 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr19_-_10697895 | 0.01 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr9_+_125273081 | 0.01 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr11_-_33744256 | 0.01 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr19_-_10687907 | 0.01 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr4_-_141348763 | 0.01 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr2_-_101925055 | 0.01 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr2_+_10091815 | 0.01 |
ENST00000324907.9
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr12_+_57482877 | 0.01 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr14_-_21905424 | 0.01 |
ENST00000553622.1
|
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr3_+_50316458 | 0.01 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr1_-_62190793 | 0.01 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr11_-_559377 | 0.01 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr11_-_62607036 | 0.01 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr1_-_32229523 | 0.01 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr1_-_53387386 | 0.01 |
ENST00000467988.1
ENST00000358358.5 ENST00000371522.4 |
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr17_+_42836521 | 0.01 |
ENST00000535346.1
|
ADAM11
|
ADAM metallopeptidase domain 11 |
| chr4_+_83956312 | 0.01 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr17_-_55038375 | 0.01 |
ENST00000240316.4
|
COIL
|
coilin |
| chr7_-_752577 | 0.01 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr9_+_116263778 | 0.01 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_+_27346666 | 0.01 |
ENST00000316470.4
ENST00000416071.1 |
ABHD1
|
abhydrolase domain containing 1 |
| chr12_+_100867486 | 0.01 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_74486109 | 0.01 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_-_13461807 | 0.01 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr16_+_29984962 | 0.01 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr9_-_131644306 | 0.01 |
ENST00000302586.3
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr1_-_25256368 | 0.01 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr9_-_131644202 | 0.01 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chrX_-_153599578 | 0.01 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr7_+_128470431 | 0.01 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr16_-_88752889 | 0.01 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr14_-_51561784 | 0.01 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr9_-_130487143 | 0.01 |
ENST00000419060.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr3_+_50192457 | 0.01 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_47021168 | 0.01 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr12_+_100867733 | 0.01 |
ENST00000546380.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_-_110052302 | 0.01 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr9_+_19049372 | 0.01 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr12_+_56401268 | 0.01 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_+_100867694 | 0.01 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_+_86396265 | 0.01 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr13_+_37574678 | 0.01 |
ENST00000389704.3
|
EXOSC8
|
exosome component 8 |
| chr19_+_5904866 | 0.01 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr19_-_46285646 | 0.01 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr16_-_70713928 | 0.01 |
ENST00000576338.1
|
MTSS1L
|
metastasis suppressor 1-like |
| chr20_-_36661826 | 0.01 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr4_-_54457783 | 0.01 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr16_-_188600 | 0.01 |
ENST00000399951.3
|
NPRL3
|
nitrogen permease regulator-like 3 (S. cerevisiae) |
| chr17_+_40925454 | 0.01 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr1_+_174846570 | 0.01 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr16_-_188624 | 0.01 |
ENST00000399953.3
|
NPRL3
|
nitrogen permease regulator-like 3 (S. cerevisiae) |
| chr19_-_40791211 | 0.01 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_41132410 | 0.01 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
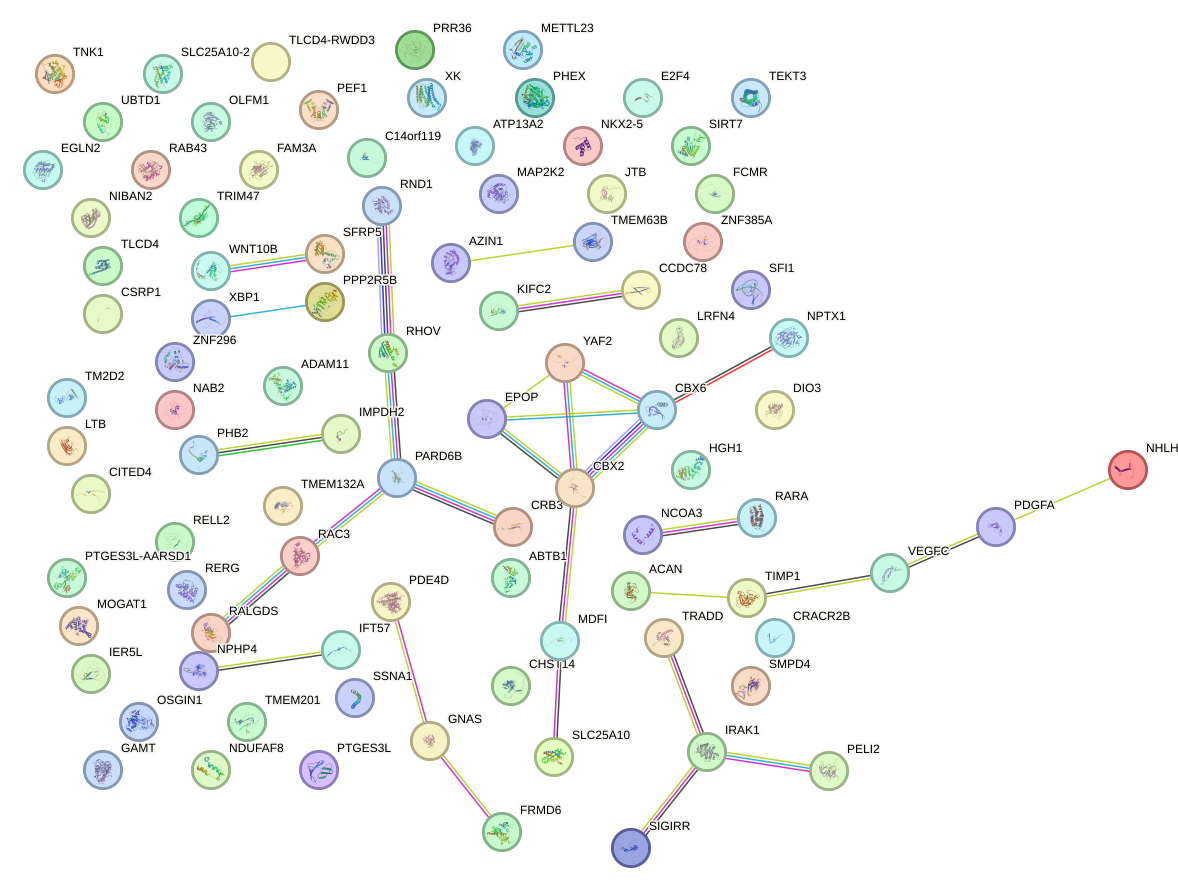
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060927 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0051885 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of anagen(GO:0051885) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |