Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HOXB5
Z-value: 0.61
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | homeobox B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg19_v2_chr17_-_46671323_46671323 | 0.32 | 6.8e-01 | Click! |
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_5322561 | 0.55 |
ENST00000396872.3
ENST00000444741.1 ENST00000297195.4 ENST00000406453.3 |
SLC29A4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr16_-_1821496 | 0.32 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr12_-_52715179 | 0.30 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr9_-_130700080 | 0.24 |
ENST00000373110.4
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr10_-_103603568 | 0.22 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr10_-_103603523 | 0.21 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr6_+_30749649 | 0.20 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr1_+_146373546 | 0.16 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr16_-_1821721 | 0.15 |
ENST00000219302.3
|
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr16_-_9770700 | 0.15 |
ENST00000561538.1
|
RP11-297M9.1
|
Uncharacterized protein |
| chrX_+_47077680 | 0.15 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr17_+_46970178 | 0.14 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chrX_+_48380205 | 0.13 |
ENST00000446158.1
ENST00000414061.1 |
EBP
|
emopamil binding protein (sterol isomerase) |
| chr19_+_7571968 | 0.11 |
ENST00000599312.1
|
CTD-2207O23.12
|
Uncharacterized protein |
| chr2_-_70409953 | 0.10 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr1_+_155579979 | 0.09 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr5_-_141061759 | 0.09 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr19_-_12267524 | 0.09 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr12_+_53693466 | 0.08 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr1_+_154229547 | 0.08 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr18_+_32558380 | 0.08 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_+_74218787 | 0.08 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr1_-_155658085 | 0.08 |
ENST00000311573.5
ENST00000438245.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr11_-_57089774 | 0.08 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr19_-_42724261 | 0.08 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr14_+_38660185 | 0.07 |
ENST00000555655.1
|
CTD-2142D14.1
|
CTD-2142D14.1 |
| chr16_-_28936007 | 0.07 |
ENST00000568703.1
ENST00000567483.1 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr7_+_125557914 | 0.07 |
ENST00000411856.1
|
AC005276.1
|
AC005276.1 |
| chr1_-_155658260 | 0.07 |
ENST00000368339.5
ENST00000405763.3 ENST00000368340.5 ENST00000454523.1 ENST00000443231.1 ENST00000347088.5 ENST00000361831.5 ENST00000355499.4 |
YY1AP1
|
YY1 associated protein 1 |
| chr7_+_120969045 | 0.07 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr2_-_74726710 | 0.07 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr16_-_30381580 | 0.07 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr6_-_133119668 | 0.07 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr12_-_6798523 | 0.07 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr19_-_16045220 | 0.06 |
ENST00000326742.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr18_-_33077556 | 0.06 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr4_+_159122728 | 0.06 |
ENST00000505049.1
ENST00000505189.1 ENST00000511038.1 |
TMEM144
|
transmembrane protein 144 |
| chr6_-_31509714 | 0.06 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr1_+_145576007 | 0.06 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr6_+_167525277 | 0.05 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr6_-_31510181 | 0.05 |
ENST00000458640.1
ENST00000396172.1 ENST00000417556.2 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chrX_+_47077632 | 0.05 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr12_+_66217911 | 0.05 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr5_-_141061777 | 0.05 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr1_+_41157671 | 0.05 |
ENST00000534399.1
ENST00000372653.1 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr1_+_155658849 | 0.05 |
ENST00000368336.5
ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3
|
death associated protein 3 |
| chr16_-_84651647 | 0.05 |
ENST00000564057.1
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chrX_+_135230712 | 0.05 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_145575980 | 0.04 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr1_-_155658518 | 0.04 |
ENST00000404643.1
ENST00000359205.5 ENST00000407221.1 |
YY1AP1
|
YY1 associated protein 1 |
| chr6_-_32820529 | 0.04 |
ENST00000425148.2
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_57089671 | 0.04 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr12_-_6798616 | 0.04 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr4_-_114900831 | 0.04 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr1_-_155658766 | 0.03 |
ENST00000295566.4
ENST00000368330.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr12_+_57984943 | 0.03 |
ENST00000422156.3
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr1_+_41157421 | 0.03 |
ENST00000372654.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr5_+_175298573 | 0.03 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr3_-_195538728 | 0.03 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr6_-_29395509 | 0.02 |
ENST00000377147.2
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chr20_-_49308048 | 0.02 |
ENST00000327979.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr6_+_121756809 | 0.02 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr17_+_2496971 | 0.02 |
ENST00000397195.5
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr17_-_29233769 | 0.02 |
ENST00000581216.1
|
TEFM
|
transcription elongation factor, mitochondrial |
| chr12_-_6798410 | 0.02 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_56101647 | 0.02 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chr2_+_219283815 | 0.02 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr3_-_164914640 | 0.02 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr12_-_53614155 | 0.02 |
ENST00000543726.1
|
RARG
|
retinoic acid receptor, gamma |
| chr1_+_116915855 | 0.02 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_+_41157361 | 0.02 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_-_17140436 | 0.01 |
ENST00000285071.4
ENST00000389169.5 ENST00000417064.1 |
FLCN
|
folliculin |
| chr14_-_24020858 | 0.01 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr1_-_248802559 | 0.01 |
ENST00000317450.3
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35 |
| chr6_-_31509506 | 0.01 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr12_-_53614043 | 0.01 |
ENST00000338561.5
|
RARG
|
retinoic acid receptor, gamma |
| chr1_+_248616077 | 0.01 |
ENST00000342927.3
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2 |
| chr17_-_42143963 | 0.01 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr14_-_102704783 | 0.01 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr1_-_119530428 | 0.01 |
ENST00000369429.3
|
TBX15
|
T-box 15 |
| chr5_+_133842243 | 0.01 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr2_+_17721920 | 0.01 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr1_+_162351503 | 0.01 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr17_+_46970134 | 0.00 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr6_-_29396243 | 0.00 |
ENST00000377148.1
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chr3_-_195538760 | 0.00 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr18_+_32558208 | 0.00 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
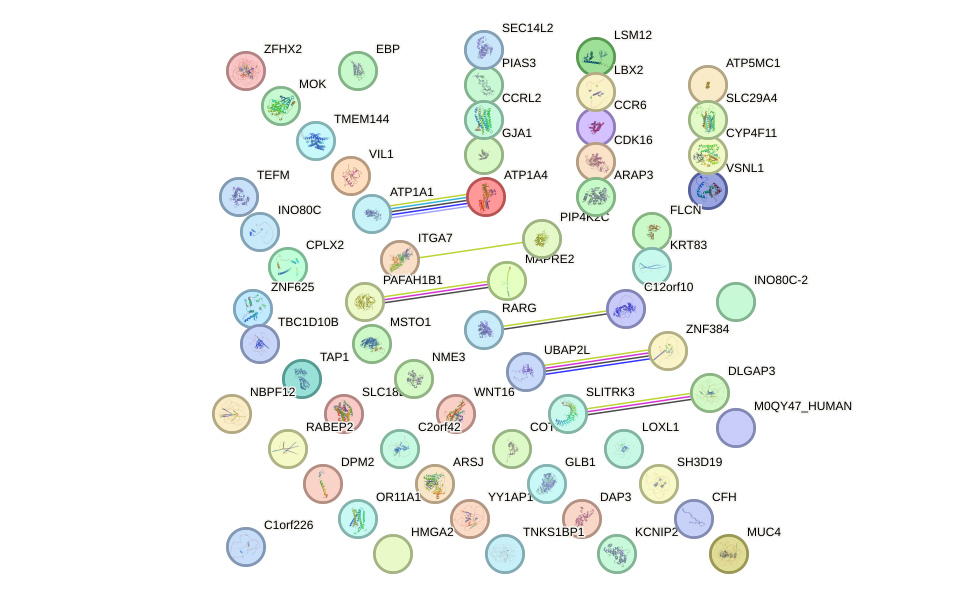
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.6 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.0 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.1 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |