Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for MAFA
Z-value: 0.31
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAF bZIP transcription factor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFA | hg19_v2_chr8_-_144512576_144512576 | 0.83 | 1.7e-01 | Click! |
Activity profile of MAFA motif
Sorted Z-values of MAFA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_58074069 | 0.25 |
ENST00000570065.1
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr7_-_150754935 | 0.11 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr12_+_21654714 | 0.11 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr4_+_74606223 | 0.10 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr15_-_66858298 | 0.10 |
ENST00000537670.1
|
LCTL
|
lactase-like |
| chr19_+_21541732 | 0.10 |
ENST00000311015.3
|
ZNF738
|
zinc finger protein 738 |
| chr6_-_74161977 | 0.08 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr22_+_22556057 | 0.07 |
ENST00000390286.2
|
IGLV11-55
|
immunoglobulin lambda variable 11-55 (non-functional) |
| chr5_-_94620239 | 0.07 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr7_-_151433342 | 0.07 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_+_36295638 | 0.07 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr11_+_777562 | 0.07 |
ENST00000530083.1
|
AP006621.5
|
Protein LOC100506518 |
| chr10_+_135122906 | 0.07 |
ENST00000368554.4
|
ZNF511
|
zinc finger protein 511 |
| chr1_+_221054411 | 0.07 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr10_-_135171178 | 0.06 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr19_-_50464338 | 0.06 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr2_+_191745560 | 0.06 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr11_+_107461948 | 0.06 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr14_+_24605389 | 0.06 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr5_-_114505624 | 0.06 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr6_+_22221010 | 0.06 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr3_+_196439170 | 0.05 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr14_+_50779029 | 0.05 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr3_+_140770183 | 0.05 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr2_+_118846008 | 0.05 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr16_-_46865047 | 0.05 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr10_+_124221036 | 0.05 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chr16_-_46865286 | 0.05 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr19_+_54641444 | 0.05 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr1_-_32403370 | 0.05 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr5_+_139739772 | 0.04 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr7_-_155160629 | 0.04 |
ENST00000378120.5
|
BLACE
|
B-cell acute lymphoblastic leukemia expressed |
| chr19_-_37958318 | 0.04 |
ENST00000316950.6
ENST00000591710.1 |
ZNF569
|
zinc finger protein 569 |
| chr19_+_12949251 | 0.04 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr2_+_103236004 | 0.04 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr16_+_28996364 | 0.04 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr5_+_138629389 | 0.04 |
ENST00000504045.1
ENST00000504311.1 ENST00000502499.1 |
MATR3
|
matrin 3 |
| chr9_+_2622085 | 0.04 |
ENST00000382099.2
|
VLDLR
|
very low density lipoprotein receptor |
| chr20_-_43743790 | 0.04 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr6_+_97372645 | 0.04 |
ENST00000536676.1
ENST00000544166.1 |
KLHL32
|
kelch-like family member 32 |
| chr6_+_7727030 | 0.04 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr3_+_196439275 | 0.04 |
ENST00000296333.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr19_+_39647271 | 0.04 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr11_+_33563821 | 0.03 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr19_-_43099070 | 0.03 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr1_-_32403903 | 0.03 |
ENST00000344035.6
ENST00000356536.3 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr13_-_28194541 | 0.03 |
ENST00000316334.3
|
LNX2
|
ligand of numb-protein X 2 |
| chr3_+_107241882 | 0.03 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chrX_-_92928557 | 0.03 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr1_-_45956868 | 0.03 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr13_-_103426081 | 0.03 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr4_+_2470664 | 0.03 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chr17_-_39781054 | 0.03 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr5_+_138629628 | 0.03 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr4_+_72052964 | 0.03 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_-_22517620 | 0.03 |
ENST00000502482.1
ENST00000334304.5 |
GPR125
|
G protein-coupled receptor 125 |
| chr5_+_138629337 | 0.03 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr16_+_56225248 | 0.03 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr11_+_118826999 | 0.03 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr20_-_23066953 | 0.03 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr6_+_42896865 | 0.03 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr4_+_72053017 | 0.03 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr17_+_41363854 | 0.03 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr15_-_23086259 | 0.03 |
ENST00000538684.1
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr6_+_97372734 | 0.03 |
ENST00000539200.1
|
KLHL32
|
kelch-like family member 32 |
| chr15_+_90744533 | 0.03 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_+_202091980 | 0.03 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr2_-_30143525 | 0.03 |
ENST00000431873.1
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr1_+_78354175 | 0.03 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr4_+_6784401 | 0.03 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr10_-_70231639 | 0.03 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr1_+_174769006 | 0.02 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr16_+_70258261 | 0.02 |
ENST00000594734.1
|
FKSG63
|
FKSG63 |
| chr4_-_10118348 | 0.02 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr8_-_63998590 | 0.02 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr7_-_30029367 | 0.02 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr3_-_196439065 | 0.02 |
ENST00000399942.4
ENST00000409690.3 |
CEP19
|
centrosomal protein 19kDa |
| chr12_+_79258444 | 0.02 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chrX_-_153141302 | 0.02 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr10_-_135171510 | 0.02 |
ENST00000278025.4
ENST00000368552.3 |
FUOM
|
fucose mutarotase |
| chr11_-_64660916 | 0.02 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr12_+_54943134 | 0.02 |
ENST00000243052.3
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent |
| chr12_+_108525517 | 0.02 |
ENST00000332082.4
|
WSCD2
|
WSC domain containing 2 |
| chr19_-_33182616 | 0.02 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr21_+_44589118 | 0.02 |
ENST00000291554.2
|
CRYAA
|
crystallin, alpha A |
| chr14_+_36295504 | 0.02 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr15_-_23086394 | 0.02 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr6_+_97372596 | 0.02 |
ENST00000369261.4
|
KLHL32
|
kelch-like family member 32 |
| chr3_-_131736593 | 0.02 |
ENST00000514999.1
|
CPNE4
|
copine IV |
| chr1_-_13331692 | 0.02 |
ENST00000353410.5
ENST00000376173.3 |
PRAMEF3
|
PRAME family member 3 |
| chr13_-_103426112 | 0.02 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr16_-_89119355 | 0.02 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr10_-_49701686 | 0.02 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr14_+_81421355 | 0.02 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr13_+_53226963 | 0.02 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr4_+_106067943 | 0.02 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr2_+_33172221 | 0.01 |
ENST00000354476.3
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr19_-_2050852 | 0.01 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr5_+_138629417 | 0.01 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr14_+_24600484 | 0.01 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr2_+_191745535 | 0.01 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr17_-_41623009 | 0.01 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr1_-_153517473 | 0.01 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr4_+_15005391 | 0.01 |
ENST00000507071.1
ENST00000345451.3 ENST00000259997.5 ENST00000382395.3 ENST00000382401.3 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr7_-_108168580 | 0.01 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr11_+_33563618 | 0.01 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr1_-_38325256 | 0.01 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr14_+_81421861 | 0.01 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr6_+_116692102 | 0.01 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr7_-_30029574 | 0.01 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr19_+_18451439 | 0.01 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr1_+_90286562 | 0.01 |
ENST00000525774.1
ENST00000337338.5 |
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr9_-_127905736 | 0.01 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr10_+_102505954 | 0.01 |
ENST00000556085.1
ENST00000427256.1 |
PAX2
|
paired box 2 |
| chr1_+_221054584 | 0.01 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr6_+_89791507 | 0.01 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_+_85095497 | 0.01 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr1_-_45956822 | 0.01 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr1_+_66458072 | 0.01 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_+_85095553 | 0.01 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr1_+_206858328 | 0.01 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr8_+_145438870 | 0.01 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr10_-_79397391 | 0.01 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_111411384 | 0.01 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chrX_+_74493896 | 0.01 |
ENST00000373383.4
ENST00000373379.1 |
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr7_-_30029328 | 0.01 |
ENST00000425819.2
ENST00000409570.1 |
SCRN1
|
secernin 1 |
| chr13_+_28194873 | 0.01 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr19_-_51141196 | 0.01 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr17_-_17109579 | 0.00 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr4_-_151936416 | 0.00 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr19_+_6740888 | 0.00 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr12_+_79258547 | 0.00 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chrX_-_153141434 | 0.00 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr16_-_67514982 | 0.00 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr11_+_111412053 | 0.00 |
ENST00000530962.1
|
LAYN
|
layilin |
| chr15_-_90456114 | 0.00 |
ENST00000398333.3
|
C15orf38-AP3S2
|
C15orf38-AP3S2 readthrough |
| chr6_+_147525541 | 0.00 |
ENST00000367481.3
ENST00000546097.1 |
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr6_-_143832820 | 0.00 |
ENST00000002165.6
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr15_+_98503922 | 0.00 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr4_-_151936865 | 0.00 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr2_-_26205550 | 0.00 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
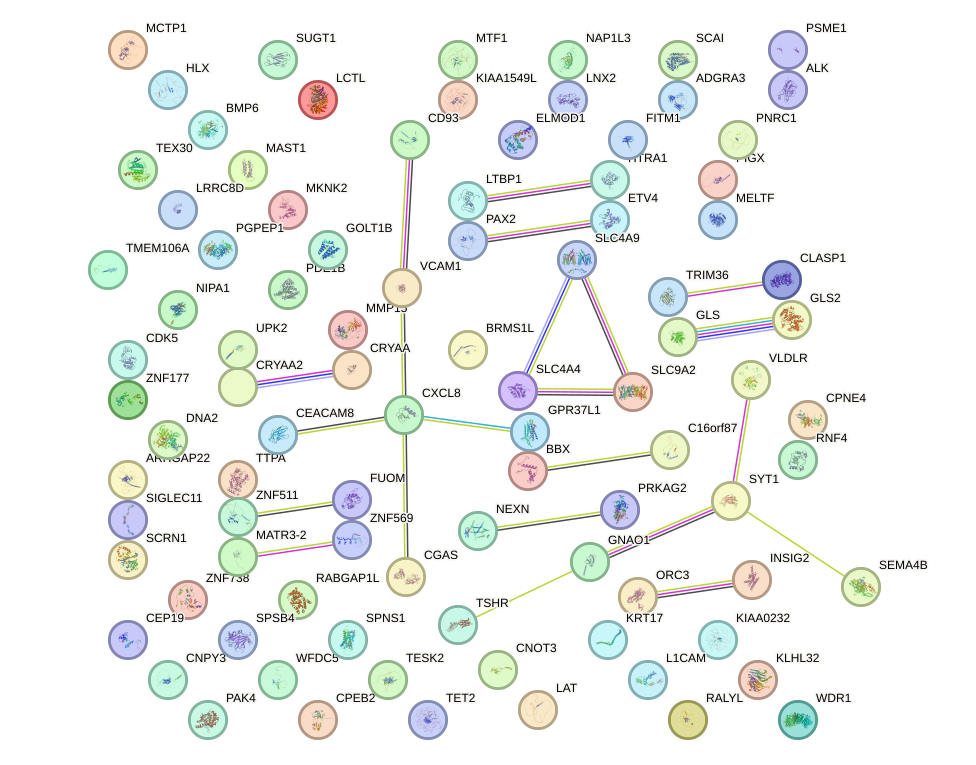
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |