Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for MBD2
Z-value: 0.56
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51751132_51751158 | -0.74 | 2.6e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_42275153 | 0.34 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr14_-_21566731 | 0.33 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr16_+_2059872 | 0.31 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr19_+_39904168 | 0.27 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_+_145137489 | 0.26 |
ENST00000355091.4
ENST00000525087.1 ENST00000361036.6 ENST00000524418.1 |
GPAA1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr3_+_112930946 | 0.25 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr19_-_41222775 | 0.24 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr5_+_149865838 | 0.24 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_-_31763276 | 0.24 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr7_+_73082152 | 0.24 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr19_-_14201507 | 0.24 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr16_+_3019309 | 0.20 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr19_-_14201776 | 0.20 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_+_56152361 | 0.20 |
ENST00000585995.1
ENST00000592996.1 |
ZNF581
CCDC106
|
zinc finger protein 581 coiled-coil domain containing 106 |
| chr12_-_49453557 | 0.18 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr16_+_3019246 | 0.17 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr15_-_90294523 | 0.17 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr2_-_220408430 | 0.16 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr11_-_64684672 | 0.16 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr2_-_175202151 | 0.16 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr4_-_170192000 | 0.16 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr10_+_104180580 | 0.16 |
ENST00000425536.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr19_-_36545649 | 0.15 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr2_+_220306238 | 0.15 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr19_-_17356697 | 0.15 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr16_-_30905584 | 0.15 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr2_+_26915584 | 0.15 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr4_+_2061119 | 0.15 |
ENST00000423729.2
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative) |
| chr16_+_2022036 | 0.15 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr17_+_36861735 | 0.14 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_-_220408260 | 0.14 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr16_+_1823208 | 0.14 |
ENST00000568449.1
ENST00000307394.7 |
EME2
|
essential meiotic structure-specific endonuclease subunit 2 |
| chr11_+_64685026 | 0.13 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr17_-_42441204 | 0.13 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr1_+_156698234 | 0.13 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr16_+_3019552 | 0.13 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr1_-_32229523 | 0.13 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr19_-_42759300 | 0.13 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr19_-_1568057 | 0.13 |
ENST00000402693.4
ENST00000388824.6 |
MEX3D
|
mex-3 RNA binding family member D |
| chr2_+_219824357 | 0.13 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr19_+_56154913 | 0.13 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr1_+_156698708 | 0.13 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr8_+_61592073 | 0.12 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr15_+_80215477 | 0.12 |
ENST00000542003.1
|
C15ORF37
|
C15orf37 protein; Uncharacterized protein |
| chr1_+_27561104 | 0.12 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr11_-_67141090 | 0.12 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr12_+_120972147 | 0.12 |
ENST00000325954.4
ENST00000542438.1 |
RNF10
|
ring finger protein 10 |
| chr12_-_120554622 | 0.12 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr12_+_57482877 | 0.11 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr12_+_122064673 | 0.11 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr12_+_6875519 | 0.11 |
ENST00000389462.4
ENST00000540874.1 ENST00000309083.6 |
PTMS
|
parathymosin |
| chr1_+_27561007 | 0.11 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr2_+_74212073 | 0.11 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr12_+_54393880 | 0.11 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr2_-_129076151 | 0.11 |
ENST00000259241.6
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr11_+_65383227 | 0.10 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr14_+_21538517 | 0.10 |
ENST00000298693.3
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr2_+_219264466 | 0.10 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr1_+_153606513 | 0.10 |
ENST00000368694.3
|
CHTOP
|
chromatin target of PRMT1 |
| chr5_-_180236811 | 0.10 |
ENST00000446023.2
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr14_+_21538429 | 0.10 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chrX_+_24167746 | 0.10 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr5_-_176778803 | 0.10 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr16_-_2059797 | 0.10 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr11_+_76092353 | 0.10 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr14_-_75593708 | 0.10 |
ENST00000557673.1
ENST00000238616.5 |
NEK9
|
NIMA-related kinase 9 |
| chr10_+_115803650 | 0.10 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr12_+_57482665 | 0.09 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_+_156698743 | 0.09 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr16_-_2059748 | 0.09 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr21_+_27543175 | 0.09 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr2_+_241375069 | 0.09 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr19_+_56152262 | 0.09 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr1_+_43855545 | 0.09 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr6_-_31763721 | 0.09 |
ENST00000375663.3
|
VARS
|
valyl-tRNA synthetase |
| chr3_-_42623805 | 0.09 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr9_+_137967395 | 0.09 |
ENST00000371801.4
|
OLFM1
|
olfactomedin 1 |
| chr19_+_14544099 | 0.09 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr12_-_50222187 | 0.09 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr11_-_75141206 | 0.09 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr1_-_27693349 | 0.09 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr19_+_36208877 | 0.08 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr20_-_30310797 | 0.08 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr2_+_11052054 | 0.08 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr14_-_99737822 | 0.08 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr22_-_50746027 | 0.08 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr2_-_128615517 | 0.08 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr1_+_153606532 | 0.08 |
ENST00000403433.1
|
CHTOP
|
chromatin target of PRMT1 |
| chr11_-_3862206 | 0.08 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr19_+_18496957 | 0.08 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr17_+_46125685 | 0.08 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr20_-_61885826 | 0.08 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr19_-_41196534 | 0.08 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr1_-_156252590 | 0.08 |
ENST00000361813.5
ENST00000368267.5 |
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr19_-_4124079 | 0.08 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr11_-_46615498 | 0.08 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr17_-_61920280 | 0.08 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr1_+_43855560 | 0.08 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr15_+_40650408 | 0.08 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr16_+_75033210 | 0.08 |
ENST00000566250.1
ENST00000567962.1 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr7_+_129710350 | 0.08 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr12_-_49182783 | 0.08 |
ENST00000550422.1
|
ADCY6
|
adenylate cyclase 6 |
| chr9_-_130616915 | 0.08 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr9_+_137967268 | 0.08 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr10_-_17659234 | 0.08 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_+_144349606 | 0.07 |
ENST00000521682.1
ENST00000340042.1 |
GLI4
|
GLI family zinc finger 4 |
| chr14_-_93581615 | 0.07 |
ENST00000555553.1
ENST00000555495.1 ENST00000554999.1 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr1_+_3689325 | 0.07 |
ENST00000444870.2
ENST00000452264.1 |
SMIM1
|
small integral membrane protein 1 (Vel blood group) |
| chr3_+_71803201 | 0.07 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr1_+_36023035 | 0.07 |
ENST00000373253.3
|
NCDN
|
neurochondrin |
| chr17_+_48638371 | 0.07 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_+_40763150 | 0.07 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr9_+_129376722 | 0.07 |
ENST00000526117.1
ENST00000373474.4 ENST00000355497.5 ENST00000425646.2 ENST00000561065.1 |
LMX1B
|
LIM homeobox transcription factor 1, beta |
| chr19_+_42388437 | 0.07 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr3_+_112930306 | 0.07 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr16_+_28303804 | 0.07 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr1_-_32229934 | 0.07 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr6_+_41514305 | 0.07 |
ENST00000409208.1
ENST00000373057.3 |
FOXP4
|
forkhead box P4 |
| chr5_+_176853669 | 0.07 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr14_+_102829381 | 0.07 |
ENST00000558678.1
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr5_+_49962495 | 0.07 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_153919128 | 0.07 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr17_+_43971643 | 0.07 |
ENST00000344290.5
ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT
|
microtubule-associated protein tau |
| chr16_-_89787360 | 0.07 |
ENST00000389386.3
|
VPS9D1
|
VPS9 domain containing 1 |
| chr9_+_35605274 | 0.07 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr17_-_80231557 | 0.07 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr17_-_79876010 | 0.07 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr10_+_75545391 | 0.07 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr12_+_122516626 | 0.07 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr3_-_52001448 | 0.07 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr11_+_67055986 | 0.07 |
ENST00000447274.2
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr11_-_62414070 | 0.06 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr19_+_38397839 | 0.06 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_-_4066890 | 0.06 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr22_-_46373004 | 0.06 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr17_+_27920486 | 0.06 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr17_+_17685422 | 0.06 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr6_+_37321748 | 0.06 |
ENST00000373479.4
ENST00000394443.4 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr11_-_67271723 | 0.06 |
ENST00000533391.1
ENST00000534749.1 ENST00000532703.1 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr7_-_19157248 | 0.06 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr11_-_75236867 | 0.06 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr9_+_135037334 | 0.06 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr20_-_48532046 | 0.06 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr3_+_33155444 | 0.06 |
ENST00000320954.6
|
CRTAP
|
cartilage associated protein |
| chr11_+_67056755 | 0.06 |
ENST00000511455.2
ENST00000308440.6 |
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr7_-_40174201 | 0.06 |
ENST00000306984.6
|
MPLKIP
|
M-phase specific PLK1 interacting protein |
| chr18_+_19750894 | 0.06 |
ENST00000581694.1
|
GATA6
|
GATA binding protein 6 |
| chr1_-_156698181 | 0.06 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr14_+_77787227 | 0.06 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr10_+_75545329 | 0.06 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr15_+_40733387 | 0.06 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr7_-_128171123 | 0.06 |
ENST00000608477.1
|
RP11-212P7.2
|
RP11-212P7.2 |
| chr14_-_77787198 | 0.06 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr9_-_135996537 | 0.06 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr17_+_74864476 | 0.06 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr17_-_31404 | 0.06 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr17_-_7142725 | 0.06 |
ENST00000571362.1
ENST00000576955.1 ENST00000320316.3 |
PHF23
|
PHD finger protein 23 |
| chr9_+_35829208 | 0.06 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr5_+_14143728 | 0.05 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr19_-_17445613 | 0.05 |
ENST00000159087.4
|
ANO8
|
anoctamin 8 |
| chr9_+_34989638 | 0.05 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr5_+_10564432 | 0.05 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr16_-_30022293 | 0.05 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr10_+_104404218 | 0.05 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr22_+_29279552 | 0.05 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr3_-_196295468 | 0.05 |
ENST00000332629.5
ENST00000433160.1 |
WDR53
|
WD repeat domain 53 |
| chr16_+_4897632 | 0.05 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr14_-_100070363 | 0.05 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr17_-_66097610 | 0.05 |
ENST00000584047.1
ENST00000579629.1 |
AC145343.2
|
AC145343.2 |
| chr9_-_130617029 | 0.05 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr11_-_6633799 | 0.05 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr8_-_142318398 | 0.05 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr11_+_118978045 | 0.05 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr2_+_176972000 | 0.05 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chrX_+_68725084 | 0.05 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr16_-_2264779 | 0.05 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr19_+_36545833 | 0.05 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr17_-_27949911 | 0.05 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr1_-_36022979 | 0.05 |
ENST00000469892.1
ENST00000325722.3 |
KIAA0319L
|
KIAA0319-like |
| chr1_-_17766198 | 0.05 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr15_+_90544532 | 0.05 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr2_+_219264762 | 0.05 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr11_+_67056867 | 0.05 |
ENST00000514166.1
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr3_+_47422485 | 0.05 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr9_+_35732312 | 0.05 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr6_+_42981922 | 0.05 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr19_+_46367518 | 0.05 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr1_+_28844778 | 0.05 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr17_-_48227877 | 0.05 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr22_-_39096661 | 0.05 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr3_-_39195037 | 0.05 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr14_+_102829300 | 0.05 |
ENST00000359520.7
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr5_+_153570319 | 0.05 |
ENST00000377661.2
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr9_-_35732362 | 0.05 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr12_+_7037461 | 0.05 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr10_-_79686284 | 0.05 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr19_+_54412517 | 0.05 |
ENST00000391767.1
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr1_-_247495045 | 0.05 |
ENST00000294753.4
ENST00000366498.2 |
ZNF496
|
zinc finger protein 496 |
| chr9_-_131644202 | 0.05 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr19_+_35491174 | 0.05 |
ENST00000317991.5
ENST00000504615.2 |
GRAMD1A
|
GRAM domain containing 1A |
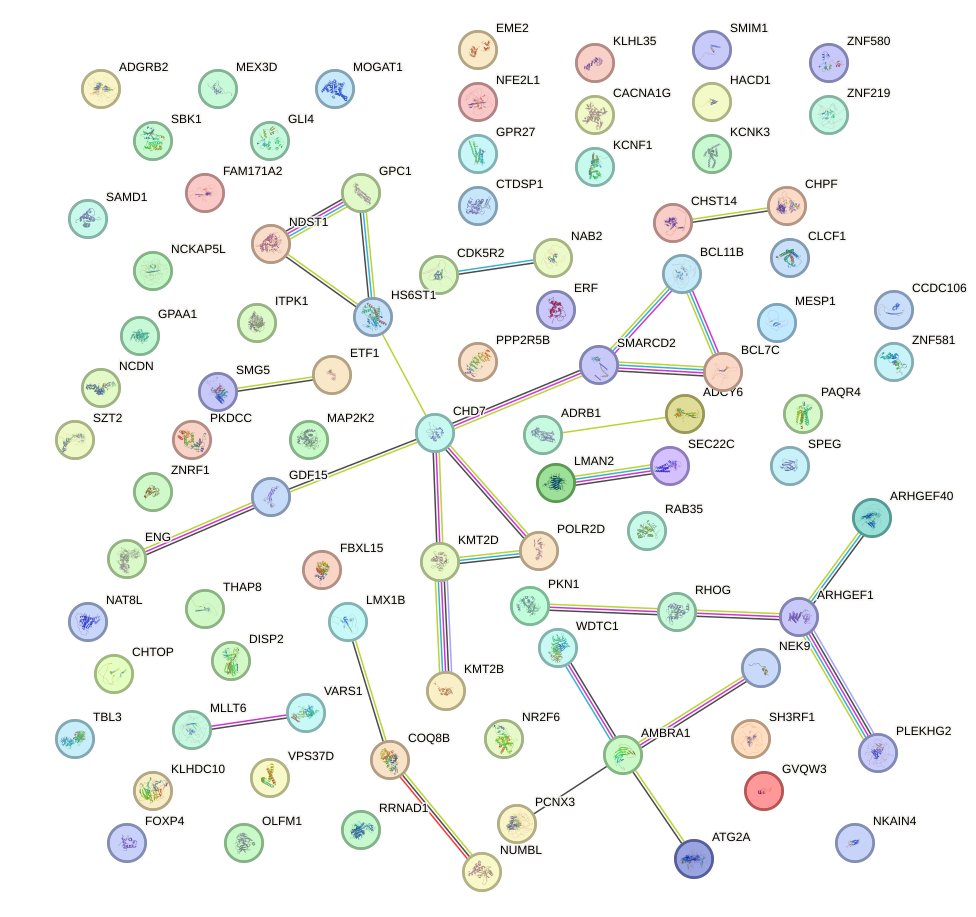
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 0.0 | GO:0060932 | bundle of His development(GO:0003166) His-Purkinje system cell differentiation(GO:0060932) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:2000275 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) regulation of oxidative phosphorylation uncoupler activity(GO:2000275) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.0 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) lung growth(GO:0060437) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0052835 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |