Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
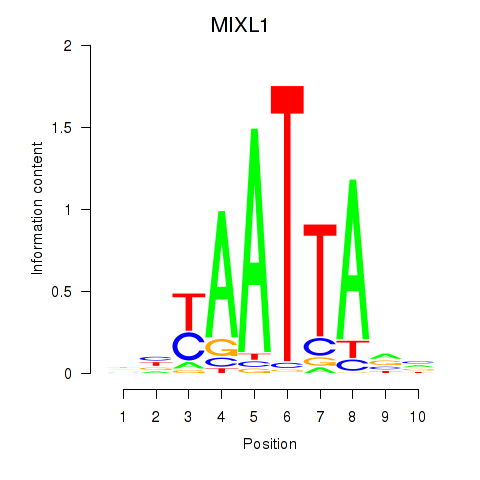
Results for MIXL1_GSX1_BSX_MEOX2_LHX4
Z-value: 0.47
Transcription factors associated with MIXL1_GSX1_BSX_MEOX2_LHX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MIXL1
|
ENSG00000185155.7 | Mix paired-like homeobox |
|
GSX1
|
ENSG00000169840.4 | GS homeobox 1 |
|
BSX
|
ENSG00000188909.4 | brain specific homeobox |
|
MEOX2
|
ENSG00000106511.5 | mesenchyme homeobox 2 |
|
LHX4
|
ENSG00000121454.4 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX4 | hg19_v2_chr1_+_180199393_180199426 | -0.42 | 5.8e-01 | Click! |
| MIXL1 | hg19_v2_chr1_+_226411319_226411366 | -0.03 | 9.7e-01 | Click! |
Activity profile of MIXL1_GSX1_BSX_MEOX2_LHX4 motif
Sorted Z-values of MIXL1_GSX1_BSX_MEOX2_LHX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_1922789 | 0.50 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr12_-_91546926 | 0.47 |
ENST00000550758.1
|
DCN
|
decorin |
| chr5_+_66300464 | 0.43 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr16_+_53412368 | 0.39 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr2_+_179318295 | 0.39 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr17_-_46690839 | 0.39 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr14_-_51027838 | 0.29 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_-_38956205 | 0.28 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr1_+_62439037 | 0.26 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr7_+_77428066 | 0.26 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr7_+_77428149 | 0.26 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_+_136070614 | 0.25 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr18_-_33709268 | 0.24 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr15_+_63188009 | 0.24 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr12_+_64798826 | 0.23 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr14_-_57960456 | 0.21 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr16_+_53133070 | 0.21 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_+_20187174 | 0.21 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr1_+_174844645 | 0.20 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_28220354 | 0.20 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr3_-_69129501 | 0.19 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr7_-_27169801 | 0.19 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr5_+_115177178 | 0.18 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_+_92114873 | 0.18 |
ENST00000343709.3
ENST00000448384.2 |
LRRC69
|
leucine rich repeat containing 69 |
| chr13_-_36788718 | 0.18 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr7_-_33102338 | 0.18 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr3_+_159557637 | 0.17 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr17_-_39623681 | 0.17 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr6_-_138833630 | 0.17 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr7_-_33102399 | 0.17 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr14_+_65016620 | 0.16 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr19_-_7698599 | 0.16 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr6_-_138866823 | 0.16 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr12_-_74686314 | 0.15 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr6_+_153552455 | 0.15 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr11_-_117747434 | 0.15 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_-_115904361 | 0.15 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr2_-_136678123 | 0.15 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr7_+_6655225 | 0.15 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr8_-_90996459 | 0.15 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr3_-_151034734 | 0.15 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr10_-_104866395 | 0.15 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr2_+_149402989 | 0.14 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_+_107334895 | 0.14 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr15_-_31393910 | 0.14 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr8_-_112248400 | 0.14 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chrX_+_13671225 | 0.14 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr12_+_16500599 | 0.14 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr15_+_80351910 | 0.14 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_+_169013666 | 0.14 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_-_114461655 | 0.14 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr1_-_242612779 | 0.14 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr5_-_142780280 | 0.14 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_39944025 | 0.13 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr13_-_44735393 | 0.13 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr10_+_13141585 | 0.13 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr12_+_104337515 | 0.13 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr6_+_126221034 | 0.13 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_-_151217166 | 0.13 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr18_-_56985873 | 0.13 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr9_-_75488984 | 0.13 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr6_+_76330355 | 0.13 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr19_-_58485895 | 0.12 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr12_+_20963647 | 0.12 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr11_-_117748138 | 0.12 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_-_55647057 | 0.12 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr4_+_146402346 | 0.12 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr14_+_101295638 | 0.12 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr10_-_105110831 | 0.12 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr2_+_109204743 | 0.11 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_123542224 | 0.11 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr17_-_57229155 | 0.11 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr13_-_41593425 | 0.11 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr12_-_112123524 | 0.11 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chrX_-_73512177 | 0.11 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chrX_-_106243451 | 0.11 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr1_-_48937838 | 0.11 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr2_-_74618907 | 0.11 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr5_+_89770664 | 0.11 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr18_+_3247779 | 0.11 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_151762943 | 0.11 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr12_+_28410128 | 0.11 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_106243294 | 0.11 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr11_-_108093329 | 0.11 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr1_+_111415757 | 0.10 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chrX_-_19817869 | 0.10 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_242612726 | 0.10 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr13_+_78315348 | 0.10 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_-_145940126 | 0.10 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr13_+_78315466 | 0.10 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_168464875 | 0.10 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr3_+_182983126 | 0.10 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr2_+_109237717 | 0.10 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_-_110937351 | 0.10 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr8_+_26150628 | 0.10 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr14_-_92247032 | 0.10 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr6_-_116833500 | 0.10 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr2_+_7118755 | 0.10 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr19_-_52307357 | 0.10 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr8_-_27469383 | 0.10 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr1_+_229440129 | 0.09 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr2_+_38152462 | 0.09 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr1_+_28527070 | 0.09 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr14_-_64194745 | 0.09 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chrX_+_149867681 | 0.09 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chrX_+_123097014 | 0.09 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr12_-_89746173 | 0.09 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr17_-_38928414 | 0.09 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr6_-_32784687 | 0.09 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr9_-_5304432 | 0.09 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr10_-_92681033 | 0.09 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_-_95391315 | 0.09 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr5_-_137475071 | 0.09 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr10_-_105110890 | 0.09 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr5_-_89770582 | 0.09 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_+_135502501 | 0.09 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr15_+_86087267 | 0.09 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr3_+_148447887 | 0.09 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr2_+_182850551 | 0.09 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr16_-_53737722 | 0.09 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr12_+_16500571 | 0.09 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr6_-_52859046 | 0.09 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_90660832 | 0.09 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr2_-_17981462 | 0.08 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr14_+_62164340 | 0.08 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr16_-_66864806 | 0.08 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr9_-_16728161 | 0.08 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr3_+_141106458 | 0.08 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr16_-_3350614 | 0.08 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr5_+_89770696 | 0.08 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr3_-_191000172 | 0.08 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr5_+_81601166 | 0.08 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr7_-_14029283 | 0.08 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr1_+_38478378 | 0.08 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr4_-_76944621 | 0.08 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr2_+_182850743 | 0.08 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_-_72070206 | 0.08 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr8_-_102216925 | 0.08 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr5_-_102455801 | 0.08 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chrX_-_46187069 | 0.08 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_+_41136144 | 0.08 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr4_-_36245561 | 0.08 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr15_+_80351977 | 0.08 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chrX_+_21874105 | 0.08 |
ENST00000429584.2
|
YY2
|
YY2 transcription factor |
| chr2_+_200472779 | 0.08 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chrX_-_71458802 | 0.08 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr3_-_108248169 | 0.08 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr18_+_32556892 | 0.08 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_-_25304889 | 0.08 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chrX_+_95939638 | 0.07 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr6_+_149887377 | 0.07 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr6_+_131571535 | 0.07 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr4_-_170897045 | 0.07 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr18_+_71815743 | 0.07 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr7_-_92747269 | 0.07 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr17_-_19619917 | 0.07 |
ENST00000325411.5
ENST00000350657.5 ENST00000433844.2 |
SLC47A2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2 |
| chr7_+_1272522 | 0.07 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr8_+_101170563 | 0.07 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr17_+_45000483 | 0.07 |
ENST00000576910.2
ENST00000439730.2 ENST00000393456.2 ENST00000415811.2 ENST00000575949.1 ENST00000225567.4 ENST00000572403.1 ENST00000570879.1 |
GOSR2
|
golgi SNAP receptor complex member 2 |
| chr9_-_21305312 | 0.07 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr19_+_52800410 | 0.07 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr11_+_107461804 | 0.07 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr12_+_16500037 | 0.07 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr19_+_56270380 | 0.07 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr1_+_112016414 | 0.07 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr18_+_57567180 | 0.07 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_+_13049413 | 0.07 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chrX_+_134654540 | 0.07 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr1_-_54411255 | 0.07 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_-_95510743 | 0.07 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr13_+_78315528 | 0.07 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr14_-_98444386 | 0.07 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr2_+_109204909 | 0.07 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr10_+_17686193 | 0.07 |
ENST00000377500.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr8_+_92114060 | 0.07 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr3_+_130569429 | 0.07 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr11_+_101918153 | 0.07 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr7_-_112635675 | 0.07 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr10_+_13141441 | 0.07 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr3_-_12587055 | 0.07 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr16_+_14280564 | 0.07 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr7_+_138145076 | 0.07 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr12_-_118628315 | 0.07 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr14_-_88200641 | 0.07 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chrX_+_70798261 | 0.07 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chrM_+_9207 | 0.07 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_+_61654271 | 0.06 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr6_+_3259148 | 0.06 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr5_-_87516448 | 0.06 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr2_+_29001711 | 0.06 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_-_44550441 | 0.06 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr12_+_122688090 | 0.06 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr3_+_111717511 | 0.06 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr12_-_54653313 | 0.06 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr8_+_38831683 | 0.06 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr6_+_28092338 | 0.06 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr4_+_183065793 | 0.06 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr4_+_76649797 | 0.06 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr17_-_38938786 | 0.06 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MIXL1_GSX1_BSX_MEOX2_LHX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.0 | GO:0016540 | protein autoprocessing(GO:0016540) positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.4 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.0 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.0 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |