Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PLAG1
Z-value: 0.33
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 zinc finger |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg19_v2_chr8_-_57123815_57123867 | 0.80 | 2.0e-01 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_12419905 | 0.20 |
ENST00000535731.1
|
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr11_+_64053311 | 0.15 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr3_+_19988736 | 0.14 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr19_-_14201776 | 0.14 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr1_-_201438282 | 0.11 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr19_-_56048456 | 0.11 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr17_+_20483037 | 0.10 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr17_+_39968926 | 0.09 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr14_-_75079294 | 0.09 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr19_-_44258770 | 0.09 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr22_-_30642782 | 0.08 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr16_-_3030283 | 0.08 |
ENST00000572619.1
ENST00000574415.1 ENST00000440027.2 ENST00000572059.1 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr17_+_38375528 | 0.08 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr16_+_30671223 | 0.08 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr19_+_56116771 | 0.08 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr16_+_29984962 | 0.08 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr16_+_4896659 | 0.07 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr19_-_44258733 | 0.07 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_-_41220957 | 0.07 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr6_+_31939608 | 0.07 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr17_-_56591321 | 0.07 |
ENST00000583243.1
|
MTMR4
|
myotubularin related protein 4 |
| chr12_-_57522813 | 0.07 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_-_1619524 | 0.07 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr17_+_48351785 | 0.07 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr6_-_33285505 | 0.07 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr2_+_220306745 | 0.06 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr7_+_75024337 | 0.06 |
ENST00000450434.1
|
TRIM73
|
tripartite motif containing 73 |
| chr7_+_73703728 | 0.06 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr1_-_151138323 | 0.06 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr19_-_42759300 | 0.06 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr19_+_40973049 | 0.06 |
ENST00000598249.1
ENST00000338932.3 ENST00000344104.3 |
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr12_+_121163538 | 0.06 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr1_-_110613276 | 0.06 |
ENST00000369792.4
|
ALX3
|
ALX homeobox 3 |
| chrX_+_153146127 | 0.06 |
ENST00000452593.1
ENST00000357566.1 |
LCA10
|
Putative lung carcinoma-associated protein 10 |
| chr17_-_4607335 | 0.06 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr17_+_48046671 | 0.06 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr19_+_39904168 | 0.05 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_-_72852320 | 0.05 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr12_+_57998400 | 0.05 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr19_-_51054299 | 0.05 |
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B |
| chr19_+_41725140 | 0.05 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr16_+_2198604 | 0.05 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr4_-_38666430 | 0.05 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr11_-_6440283 | 0.05 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr14_+_79745682 | 0.05 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr11_+_66610883 | 0.05 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr10_-_21786179 | 0.05 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr11_+_65343494 | 0.04 |
ENST00000309295.4
ENST00000533237.1 |
EHBP1L1
|
EH domain binding protein 1-like 1 |
| chr12_-_12419703 | 0.04 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr11_-_57282349 | 0.04 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr14_-_51562037 | 0.04 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr5_+_10353780 | 0.04 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr6_-_43021437 | 0.04 |
ENST00000265348.3
|
CUL7
|
cullin 7 |
| chr8_+_21912328 | 0.04 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr11_-_94964210 | 0.04 |
ENST00000416495.2
ENST00000393234.1 |
SESN3
|
sestrin 3 |
| chr10_-_75571566 | 0.04 |
ENST00000299641.4
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr3_-_52001448 | 0.04 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr10_-_102890883 | 0.04 |
ENST00000445873.1
|
TLX1NB
|
TLX1 neighbor |
| chr3_-_184971817 | 0.04 |
ENST00000440662.1
ENST00000456310.1 |
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr18_-_51750948 | 0.04 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_+_75024903 | 0.04 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr10_-_126849626 | 0.04 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_-_145470383 | 0.04 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr5_-_176936817 | 0.04 |
ENST00000502885.1
ENST00000506493.1 |
DOK3
|
docking protein 3 |
| chr19_+_18283959 | 0.04 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr1_-_115632035 | 0.04 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr16_+_31483374 | 0.04 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr6_-_31940065 | 0.04 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr17_-_79805146 | 0.04 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr16_+_31483451 | 0.04 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chrX_+_51927919 | 0.03 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr9_+_2621950 | 0.03 |
ENST00000382096.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr22_+_51112800 | 0.03 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_-_15374033 | 0.03 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chrX_-_16888276 | 0.03 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr1_-_27701307 | 0.03 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr2_-_74619152 | 0.03 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chrY_+_22737604 | 0.03 |
ENST00000361365.2
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr7_-_76039000 | 0.03 |
ENST00000275560.3
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr17_+_7462031 | 0.03 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_-_204654481 | 0.03 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chrX_+_48398053 | 0.03 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr1_+_154193643 | 0.03 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr19_+_40697514 | 0.03 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr1_-_212003556 | 0.03 |
ENST00000366996.1
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr4_-_57844989 | 0.03 |
ENST00000264230.4
|
NOA1
|
nitric oxide associated 1 |
| chr6_-_43021612 | 0.03 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr12_+_57522439 | 0.03 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr11_+_63997750 | 0.03 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr15_+_41186609 | 0.03 |
ENST00000220509.5
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr1_-_68698222 | 0.02 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr16_+_31191431 | 0.02 |
ENST00000254108.7
ENST00000380244.3 ENST00000568685.1 |
FUS
|
fused in sarcoma |
| chr3_+_14989076 | 0.02 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr9_-_123476719 | 0.02 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr11_-_73309228 | 0.02 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr22_+_29469012 | 0.02 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr12_+_105724613 | 0.02 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr20_+_35201857 | 0.02 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr15_+_90544532 | 0.02 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr12_+_58005204 | 0.02 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr6_+_31730773 | 0.02 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr10_-_75571341 | 0.02 |
ENST00000309979.6
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr11_-_73309112 | 0.02 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr12_+_57522258 | 0.02 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr1_+_202431859 | 0.02 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_56546363 | 0.02 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr22_+_19744226 | 0.02 |
ENST00000332710.4
ENST00000329705.7 ENST00000359500.3 |
TBX1
|
T-box 1 |
| chr9_+_2621798 | 0.02 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr6_-_150039170 | 0.02 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr11_-_61062762 | 0.02 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr22_+_29469100 | 0.02 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr4_+_55095428 | 0.02 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr17_-_1420006 | 0.01 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr19_+_41083064 | 0.01 |
ENST00000595631.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr9_-_136004782 | 0.01 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr3_+_49711391 | 0.01 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr15_-_41408409 | 0.01 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr16_-_29910853 | 0.01 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr15_+_29131103 | 0.01 |
ENST00000558402.1
ENST00000558330.1 |
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr19_+_45844018 | 0.01 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr9_-_123476612 | 0.01 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr12_+_79258547 | 0.01 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr19_-_14201507 | 0.01 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr10_+_24497704 | 0.01 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr1_-_153931052 | 0.01 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr2_-_85788652 | 0.01 |
ENST00000430215.3
|
GGCX
|
gamma-glutamyl carboxylase |
| chr6_+_106534192 | 0.01 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr1_-_68698197 | 0.01 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr19_+_41725088 | 0.01 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chr11_-_111170526 | 0.01 |
ENST00000355430.4
|
COLCA1
|
colorectal cancer associated 1 |
| chr10_+_102758105 | 0.01 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr6_+_43044003 | 0.01 |
ENST00000230419.4
ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7
|
protein tyrosine kinase 7 |
| chr20_+_35169885 | 0.01 |
ENST00000279022.2
ENST00000346786.2 |
MYL9
|
myosin, light chain 9, regulatory |
| chr4_-_114682936 | 0.01 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr19_+_45843994 | 0.01 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr4_+_55095264 | 0.01 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr12_-_112819896 | 0.01 |
ENST00000377560.5
ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chrX_+_152599604 | 0.01 |
ENST00000370251.3
ENST00000421401.3 |
ZNF275
|
zinc finger protein 275 |
| chr1_-_151138422 | 0.01 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr17_+_30593195 | 0.00 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr2_-_152955537 | 0.00 |
ENST00000201943.5
ENST00000539935.1 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_-_152955213 | 0.00 |
ENST00000427385.1
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr15_-_43910438 | 0.00 |
ENST00000432436.1
|
STRC
|
stereocilin |
| chr7_-_22233442 | 0.00 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr10_+_24498060 | 0.00 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr15_-_41408339 | 0.00 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chrX_+_110339439 | 0.00 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr16_+_2521500 | 0.00 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr20_+_62716348 | 0.00 |
ENST00000349451.3
|
OPRL1
|
opiate receptor-like 1 |
| chr2_-_74618964 | 0.00 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr14_-_25519095 | 0.00 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr14_-_25519317 | 0.00 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr15_+_92397051 | 0.00 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr14_+_57857262 | 0.00 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
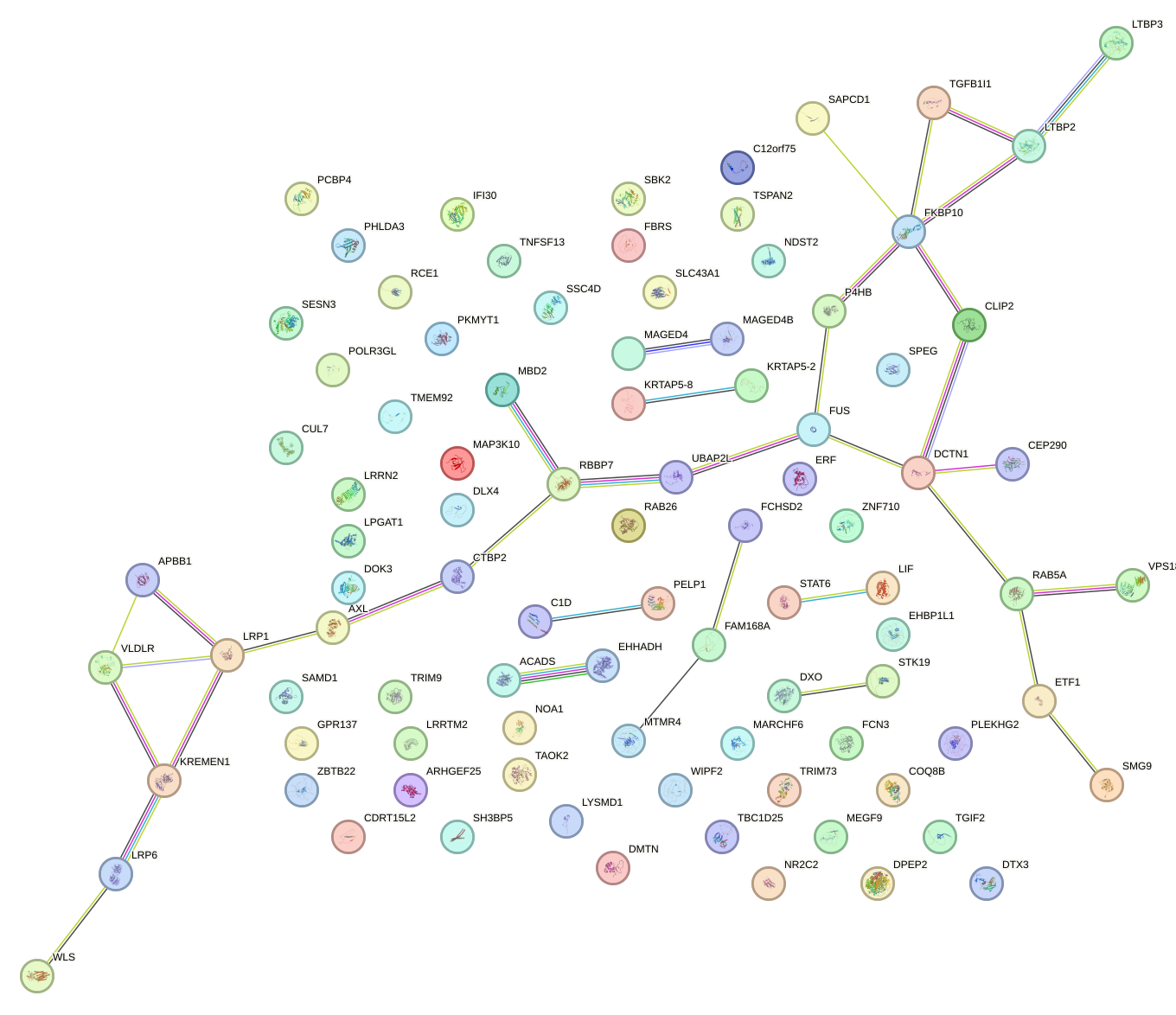
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.0 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.0 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |