Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PPARA
Z-value: 0.52
Transcription factors associated with PPARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARA
|
ENSG00000186951.12 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARA | hg19_v2_chr22_+_46546494_46546525 | -0.81 | 1.9e-01 | Click! |
Activity profile of PPARA motif
Sorted Z-values of PPARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_65337708 | 0.45 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr3_-_117716418 | 0.40 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr17_-_49021974 | 0.33 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr10_+_81065975 | 0.29 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr11_+_73498973 | 0.28 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr1_-_12679171 | 0.26 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr9_-_35685452 | 0.25 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr5_-_87564620 | 0.24 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr1_-_85156417 | 0.23 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr17_+_48046671 | 0.22 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr15_+_23810903 | 0.21 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr1_+_46640750 | 0.21 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr14_+_32476072 | 0.20 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr1_+_145727681 | 0.19 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr19_-_51568324 | 0.18 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr16_+_58535372 | 0.17 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr15_+_59730348 | 0.17 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr19_-_4535233 | 0.16 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr14_-_21492251 | 0.15 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr7_+_128399002 | 0.15 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr17_-_7123021 | 0.15 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr16_+_53133070 | 0.15 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_16705069 | 0.15 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr17_+_7533439 | 0.15 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr1_-_160231451 | 0.14 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr7_-_72936608 | 0.14 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_-_131299929 | 0.14 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_+_58765103 | 0.14 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_131028869 | 0.14 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr12_-_7281469 | 0.14 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr1_+_46769303 | 0.14 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr12_+_57854274 | 0.13 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr8_-_131028660 | 0.13 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr4_+_26322409 | 0.13 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_82555125 | 0.13 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr7_-_72936531 | 0.12 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr22_+_22676808 | 0.12 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr9_-_34665983 | 0.12 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr16_+_14280742 | 0.12 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr11_-_64013663 | 0.12 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr12_-_6715808 | 0.12 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr6_+_107349392 | 0.12 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr3_-_167452262 | 0.12 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr19_-_49371711 | 0.12 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr8_-_125486755 | 0.12 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr8_+_97597148 | 0.12 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr21_-_27107198 | 0.11 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr17_-_7082668 | 0.11 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_-_113464906 | 0.11 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr8_-_131028782 | 0.11 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr11_+_73498898 | 0.11 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr13_+_41635617 | 0.11 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr2_-_177684007 | 0.11 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr4_+_106067943 | 0.11 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr1_-_26233423 | 0.11 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr14_-_23791484 | 0.11 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr12_-_96793142 | 0.11 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr20_-_35807741 | 0.11 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr3_-_167813132 | 0.11 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr7_-_140624499 | 0.10 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr3_-_42845922 | 0.10 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr12_-_16760195 | 0.10 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_-_72129806 | 0.10 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr2_-_25194476 | 0.10 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr1_+_32666188 | 0.10 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr3_-_167452298 | 0.10 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr5_-_95550754 | 0.10 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr1_+_63788730 | 0.10 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr8_-_53626974 | 0.10 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr8_+_110346546 | 0.10 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr14_+_21566980 | 0.10 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr3_-_45957088 | 0.10 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_204183071 | 0.10 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr12_-_95397442 | 0.10 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr10_+_51371390 | 0.10 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chrX_-_108868390 | 0.10 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr14_+_58765305 | 0.10 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr5_-_60139347 | 0.10 |
ENST00000511799.1
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr15_+_58724184 | 0.10 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr1_+_29213678 | 0.10 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr22_+_23248512 | 0.10 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr3_-_45957534 | 0.10 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_205719295 | 0.09 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr1_+_45140400 | 0.09 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr7_-_105319536 | 0.09 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr8_-_94928861 | 0.09 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2 |
| chr5_+_140027355 | 0.09 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr14_+_61654271 | 0.09 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr2_+_33701707 | 0.09 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr20_-_13765526 | 0.09 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr4_-_668108 | 0.09 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr21_-_27107344 | 0.09 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_-_91487770 | 0.09 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr3_-_122102065 | 0.09 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chr21_-_27107283 | 0.09 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_-_131211534 | 0.09 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_+_26986582 | 0.09 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr9_-_34381536 | 0.09 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chrX_-_46187069 | 0.09 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr15_+_71145578 | 0.09 |
ENST00000544974.2
ENST00000558546.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_17380630 | 0.09 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr6_+_31939608 | 0.09 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr22_+_29469100 | 0.09 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr1_-_91487806 | 0.08 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr5_+_118406796 | 0.08 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr14_+_21492331 | 0.08 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr1_-_211848899 | 0.08 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr10_+_76586348 | 0.08 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr2_+_201936458 | 0.08 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr10_+_81107271 | 0.08 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr15_+_35838396 | 0.08 |
ENST00000501169.2
|
DPH6-AS1
|
DPH6 antisense RNA 1 (head to head) |
| chr2_-_198364581 | 0.08 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr14_-_50053081 | 0.08 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr3_-_52002194 | 0.08 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr11_+_111169565 | 0.08 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr16_+_85942594 | 0.08 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr3_+_37284668 | 0.08 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr10_-_94050820 | 0.08 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr19_+_16186903 | 0.08 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr16_-_4588469 | 0.08 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr22_-_38699003 | 0.08 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr14_+_23791159 | 0.08 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr4_+_146560245 | 0.08 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_117220016 | 0.08 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr1_+_68150744 | 0.08 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_+_23810853 | 0.08 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr19_+_1407733 | 0.08 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr7_+_121513143 | 0.08 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr2_-_165698662 | 0.08 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_219433014 | 0.08 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr5_+_172332220 | 0.08 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr8_+_66955648 | 0.08 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr20_-_30458354 | 0.08 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr14_-_92572894 | 0.08 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr1_+_45140360 | 0.08 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr7_-_83824449 | 0.07 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_+_50794947 | 0.07 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr14_-_31889737 | 0.07 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr14_+_75761099 | 0.07 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr7_-_123198284 | 0.07 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr3_-_141747950 | 0.07 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_156675564 | 0.07 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr8_+_94929273 | 0.07 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_+_218356 | 0.07 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr14_-_104387888 | 0.07 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr12_-_48744554 | 0.07 |
ENST00000544117.2
ENST00000548932.1 ENST00000549125.1 ENST00000301042.3 ENST00000547026.1 |
ZNF641
|
zinc finger protein 641 |
| chr10_-_65028817 | 0.07 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr7_+_79764104 | 0.07 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr9_-_88874519 | 0.07 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr17_+_7517264 | 0.07 |
ENST00000593717.1
ENST00000572182.1 ENST00000574539.1 ENST00000576728.1 ENST00000575314.1 ENST00000570547.1 ENST00000572262.1 ENST00000576478.1 |
AC007421.1
SHBG
|
AC007421.1 sex hormone-binding globulin |
| chr1_+_33439268 | 0.07 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr12_-_111926342 | 0.07 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr14_-_24729251 | 0.07 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr14_+_73525229 | 0.07 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr3_-_197024965 | 0.07 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_67563250 | 0.07 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr19_+_1407517 | 0.07 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr3_+_113667354 | 0.07 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr4_+_26322185 | 0.07 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_-_151433393 | 0.07 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr16_-_68269971 | 0.07 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chrX_-_109683446 | 0.07 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr3_-_71114066 | 0.07 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr19_-_51289436 | 0.07 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr2_+_230787213 | 0.07 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr8_-_80942139 | 0.07 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr2_+_219745020 | 0.07 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr12_+_100897130 | 0.07 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr8_-_100905925 | 0.07 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr3_+_14989186 | 0.07 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr5_+_162887556 | 0.07 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr8_-_131028641 | 0.07 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr9_+_103189660 | 0.07 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr3_+_159943362 | 0.07 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_+_32006042 | 0.07 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr1_+_222988406 | 0.07 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr5_-_79287060 | 0.07 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr6_-_149969829 | 0.07 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr11_+_64008443 | 0.07 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_213224572 | 0.07 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr6_+_149068464 | 0.07 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr19_+_6739662 | 0.07 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr8_+_63161491 | 0.07 |
ENST00000523211.1
ENST00000524201.1 |
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr14_+_69951457 | 0.07 |
ENST00000322564.7
|
PLEKHD1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr7_-_151433342 | 0.07 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr3_-_52486841 | 0.07 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr11_+_64008525 | 0.07 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_162016827 | 0.07 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_82555169 | 0.06 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr4_-_143767428 | 0.06 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_-_91875358 | 0.06 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr4_+_159593418 | 0.06 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_+_27849378 | 0.06 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr14_+_71788096 | 0.06 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr2_+_114256661 | 0.06 |
ENST00000306507.5
|
FOXD4L1
|
forkhead box D4-like 1 |
| chr4_-_100815525 | 0.06 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr12_-_6483969 | 0.06 |
ENST00000396966.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr2_-_44223089 | 0.06 |
ENST00000447246.1
ENST00000409946.1 ENST00000409659.1 |
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr15_-_75017711 | 0.06 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr19_-_49552363 | 0.06 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
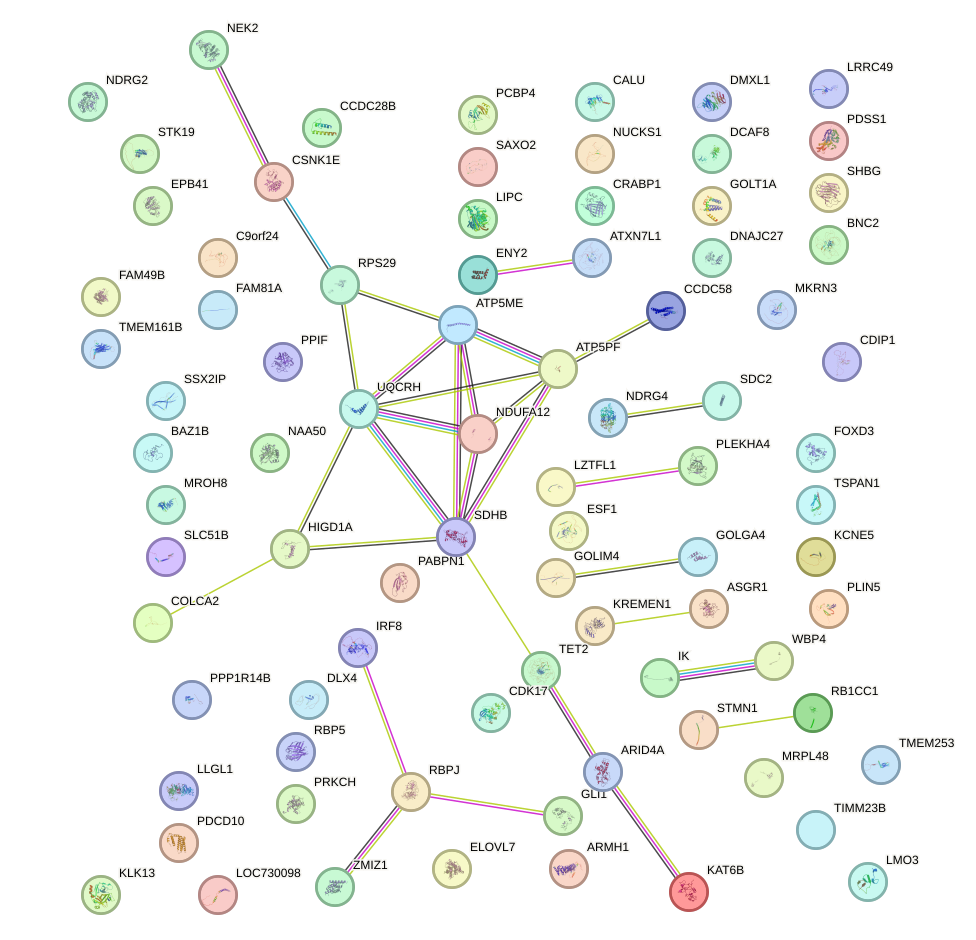
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.5 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.3 | GO:0072554 | blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.1 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.0 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |