Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SIX1_SIX3_SIX2
Z-value: 0.28
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | -0.86 | 1.4e-01 | Click! |
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.81 | 1.9e-01 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.54 | 4.6e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_139025875 | 0.18 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr1_-_44818599 | 0.17 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr16_+_777739 | 0.15 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr20_+_34824355 | 0.14 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr6_-_154751629 | 0.12 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr19_+_50015870 | 0.12 |
ENST00000599701.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr13_-_30160925 | 0.12 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr7_-_48068699 | 0.11 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr19_+_50270219 | 0.11 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr15_+_40650408 | 0.11 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr7_+_150759634 | 0.10 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr15_-_31521567 | 0.09 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr11_+_63742050 | 0.08 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr15_+_75491203 | 0.08 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr6_+_26273144 | 0.08 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr7_-_34978980 | 0.08 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr9_-_27005686 | 0.08 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr1_-_108743471 | 0.07 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_+_100551239 | 0.07 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr14_+_51026844 | 0.07 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr11_-_2924720 | 0.07 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr22_+_19710468 | 0.07 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr11_-_44972418 | 0.07 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr13_+_28519343 | 0.07 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr7_+_99746514 | 0.07 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr11_+_73661364 | 0.07 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr14_-_24911448 | 0.06 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr20_+_62694834 | 0.06 |
ENST00000415602.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr19_+_14551066 | 0.06 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr1_-_149982624 | 0.06 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr12_+_7052974 | 0.06 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr12_+_110011571 | 0.06 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr19_+_11457175 | 0.06 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr3_+_149192475 | 0.06 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr6_-_32160622 | 0.06 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr5_+_125758813 | 0.06 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_158656488 | 0.06 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr10_-_103603523 | 0.06 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr19_+_56116771 | 0.06 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr12_-_111395610 | 0.06 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chr15_+_91416092 | 0.05 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_+_5238793 | 0.05 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr12_+_57624085 | 0.05 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_-_11457162 | 0.05 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr17_+_7465216 | 0.05 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr11_-_503521 | 0.05 |
ENST00000534797.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr11_-_62559455 | 0.05 |
ENST00000528367.1
ENST00000525631.1 ENST00000307366.7 |
TMEM223
|
transmembrane protein 223 |
| chr5_+_125758865 | 0.05 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_+_114168773 | 0.05 |
ENST00000542647.1
ENST00000545255.1 |
NNMT
|
nicotinamide N-methyltransferase |
| chr16_+_29832634 | 0.05 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr4_-_860950 | 0.05 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr19_-_55549624 | 0.05 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr16_+_2303738 | 0.05 |
ENST00000454671.1
|
AC009065.1
|
Uncharacterized protein |
| chr8_-_145652336 | 0.05 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr7_+_75511362 | 0.05 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr1_-_1310870 | 0.05 |
ENST00000338338.5
|
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr9_+_131549610 | 0.05 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr12_-_58220078 | 0.05 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr16_-_28550320 | 0.05 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr7_-_1609591 | 0.04 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr12_+_70574088 | 0.04 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr15_+_75491213 | 0.04 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr14_-_74296806 | 0.04 |
ENST00000555539.1
|
RP5-1021I20.2
|
RP5-1021I20.2 |
| chr1_+_241695670 | 0.04 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_+_65999265 | 0.04 |
ENST00000528935.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr11_-_44972476 | 0.04 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr17_+_46184911 | 0.04 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr2_+_217498105 | 0.04 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_-_31763721 | 0.04 |
ENST00000375663.3
|
VARS
|
valyl-tRNA synthetase |
| chr1_+_186265399 | 0.04 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr3_+_149191723 | 0.04 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_-_50836762 | 0.04 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| chr16_-_2581409 | 0.04 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
| chr19_+_49109990 | 0.04 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr22_-_43091639 | 0.04 |
ENST00000249005.2
ENST00000381278.3 |
A4GALT
|
alpha 1,4-galactosyltransferase |
| chr20_-_33732952 | 0.04 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr4_+_89300158 | 0.04 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_+_57623869 | 0.04 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr4_+_129349188 | 0.04 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr1_-_8000872 | 0.04 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_+_57624119 | 0.04 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_156698708 | 0.04 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_-_61517572 | 0.04 |
ENST00000582997.1
|
CYB561
|
cytochrome b561 |
| chr19_-_3600549 | 0.04 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr2_-_150444116 | 0.04 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr16_+_68279256 | 0.04 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr11_+_114168085 | 0.04 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr12_+_56862301 | 0.04 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr19_-_50316517 | 0.04 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr10_-_103603568 | 0.04 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr16_+_451826 | 0.04 |
ENST00000219481.5
ENST00000397710.1 ENST00000424398.2 |
DECR2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr19_+_36605850 | 0.04 |
ENST00000221855.3
|
TBCB
|
tubulin folding cofactor B |
| chr1_+_241695424 | 0.03 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_8579030 | 0.03 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr1_-_12908578 | 0.03 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr1_+_156698743 | 0.03 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr6_-_31509714 | 0.03 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr19_+_17579556 | 0.03 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr19_-_11039261 | 0.03 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr9_+_131549483 | 0.03 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr11_-_59633951 | 0.03 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr12_-_11036844 | 0.03 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr17_-_33446735 | 0.03 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr2_-_220034745 | 0.03 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr12_+_57623907 | 0.03 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_-_207119738 | 0.03 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr20_+_44637526 | 0.03 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr14_-_24740709 | 0.03 |
ENST00000399409.3
ENST00000216840.6 |
RABGGTA
|
Rab geranylgeranyltransferase, alpha subunit |
| chr19_-_52097613 | 0.03 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr11_-_44972390 | 0.03 |
ENST00000395648.3
ENST00000531928.2 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr21_+_27011584 | 0.03 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr2_+_208414985 | 0.03 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr16_+_31119615 | 0.03 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr10_-_103603677 | 0.03 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr2_+_161993465 | 0.03 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_-_31671058 | 0.03 |
ENST00000538874.1
ENST00000395952.3 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr8_+_99129513 | 0.03 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr19_+_45445491 | 0.03 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr16_+_56642041 | 0.03 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chrX_-_72095808 | 0.03 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
| chr4_+_89514402 | 0.03 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_-_179457805 | 0.03 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr17_-_45056606 | 0.03 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr5_+_125759140 | 0.03 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr10_+_135207623 | 0.03 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr19_+_54369434 | 0.03 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr12_-_7077125 | 0.03 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr8_-_48651648 | 0.03 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr12_+_113796347 | 0.03 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr15_+_62853562 | 0.03 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr14_+_61449076 | 0.03 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr20_-_44600810 | 0.03 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr19_-_58446721 | 0.03 |
ENST00000396147.1
ENST00000595569.1 ENST00000599852.1 ENST00000425570.3 ENST00000601593.1 |
ZNF418
|
zinc finger protein 418 |
| chr11_+_100862811 | 0.03 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr16_+_29802036 | 0.03 |
ENST00000561482.1
ENST00000160827.4 ENST00000569636.2 ENST00000400750.2 |
KIF22
|
kinesin family member 22 |
| chr3_-_111314230 | 0.03 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr16_-_28550348 | 0.03 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr6_+_160693591 | 0.03 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr8_+_35649365 | 0.03 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr9_+_6716478 | 0.03 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr12_-_49523896 | 0.03 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr2_-_150444300 | 0.03 |
ENST00000303319.5
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr20_-_36661826 | 0.03 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr19_-_46146946 | 0.03 |
ENST00000536630.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr22_-_44258360 | 0.02 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr1_+_113010056 | 0.02 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr18_-_53257027 | 0.02 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr15_-_43212996 | 0.02 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr12_+_57624059 | 0.02 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_54718904 | 0.02 |
ENST00000262061.2
ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr11_-_125932685 | 0.02 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr10_+_5005598 | 0.02 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr1_+_196743912 | 0.02 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr17_-_41466555 | 0.02 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr20_+_814377 | 0.02 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr2_+_62132800 | 0.02 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr14_+_91709103 | 0.02 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr19_-_39226045 | 0.02 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr2_-_9563319 | 0.02 |
ENST00000497105.1
ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr17_+_6915798 | 0.02 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr7_+_72349920 | 0.02 |
ENST00000395270.1
ENST00000446813.1 ENST00000257622.4 |
POM121
|
POM121 transmembrane nucleoporin |
| chr17_-_4938712 | 0.02 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr11_-_125365435 | 0.02 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr19_+_11039391 | 0.02 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr16_+_28763108 | 0.02 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr1_+_198126209 | 0.02 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr12_-_58135903 | 0.02 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_+_5005445 | 0.02 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr19_-_48389651 | 0.02 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr12_-_53901266 | 0.02 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr3_-_178976996 | 0.02 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr9_+_131062367 | 0.02 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr19_+_47523058 | 0.02 |
ENST00000602212.1
ENST00000602189.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr10_+_135207598 | 0.02 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr15_-_55562582 | 0.02 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_197237352 | 0.02 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr19_-_1848451 | 0.02 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr19_+_1026298 | 0.02 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr12_-_118490217 | 0.02 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr3_+_42850959 | 0.02 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr3_+_196669494 | 0.02 |
ENST00000602845.1
|
NCBP2-AS2
|
NCBP2 antisense RNA 2 (head to head) |
| chr17_+_6915730 | 0.02 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr11_-_130786333 | 0.02 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr15_-_43212836 | 0.02 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr19_+_11750566 | 0.02 |
ENST00000344893.3
|
ZNF833P
|
zinc finger protein 833, pseudogene |
| chr11_+_61520075 | 0.02 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr2_+_219135115 | 0.02 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr10_-_49860525 | 0.02 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_-_158732340 | 0.02 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr19_+_17862274 | 0.02 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr7_+_870547 | 0.02 |
ENST00000457598.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr17_+_6915902 | 0.02 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr11_+_102552041 | 0.02 |
ENST00000537079.1
|
RP11-817J15.3
|
Uncharacterized protein |
| chr19_-_11039188 | 0.02 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr6_-_31509506 | 0.02 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr8_+_119294456 | 0.02 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr1_-_51791596 | 0.02 |
ENST00000532836.1
ENST00000422925.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr19_-_41942344 | 0.02 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
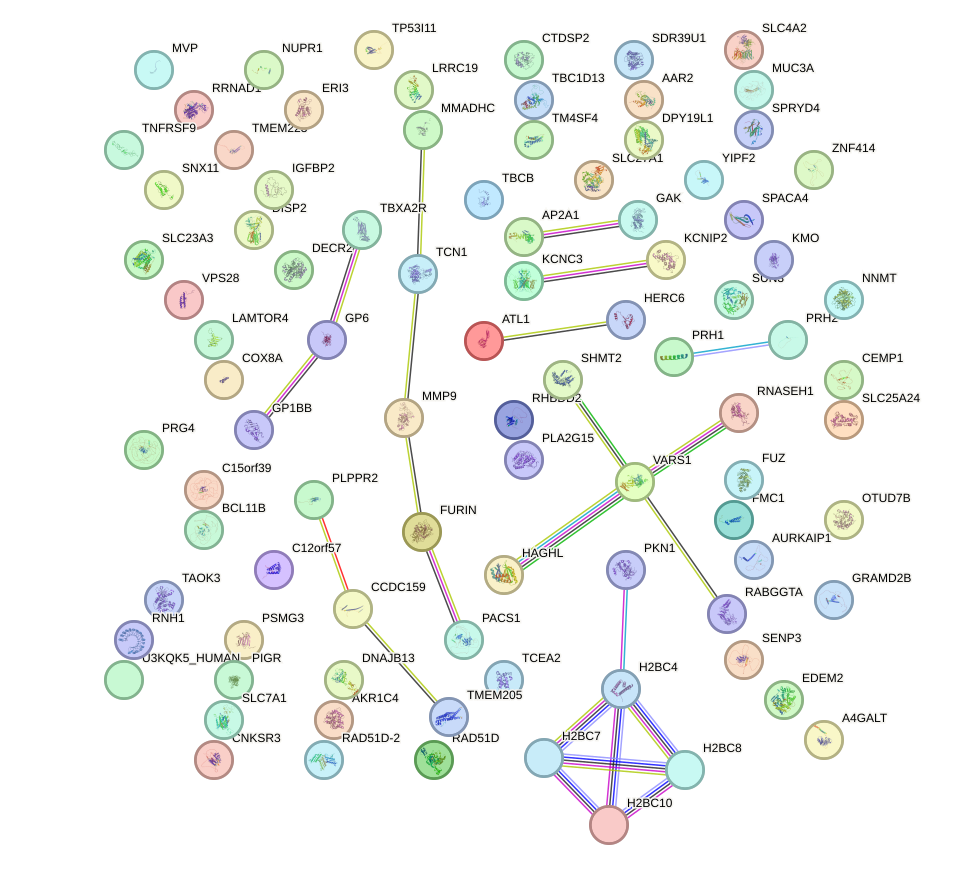
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.0 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |