Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SOX1
Z-value: 0.38
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_138866823 | 0.08 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr8_-_99955042 | 0.08 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr7_-_139756791 | 0.08 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr6_+_148593425 | 0.08 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chrX_-_11129229 | 0.07 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr19_-_6057282 | 0.07 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr3_-_107596910 | 0.07 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr5_-_96209315 | 0.07 |
ENST00000504056.1
|
CTD-2260A17.2
|
Uncharacterized protein |
| chr2_-_163175133 | 0.07 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr8_-_134501937 | 0.07 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr3_-_58200398 | 0.06 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr4_-_74486109 | 0.06 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr20_-_49575081 | 0.05 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr16_-_18937072 | 0.05 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr3_+_138327417 | 0.05 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_+_27833034 | 0.05 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr20_-_49575058 | 0.05 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_+_136676707 | 0.05 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr8_-_134501873 | 0.05 |
ENST00000523634.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr16_+_12059091 | 0.05 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr11_-_6790286 | 0.04 |
ENST00000338569.2
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2 |
| chr4_-_74486217 | 0.04 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_138327542 | 0.04 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr10_-_73976884 | 0.04 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr11_-_113577014 | 0.04 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr10_-_73976025 | 0.04 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr15_+_42565393 | 0.03 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr9_-_95244781 | 0.03 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr21_-_16126181 | 0.03 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr4_-_170924888 | 0.03 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_+_172734548 | 0.03 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chrX_-_57021943 | 0.03 |
ENST00000374919.3
|
SPIN3
|
spindlin family, member 3 |
| chr17_-_38821373 | 0.03 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_184020811 | 0.03 |
ENST00000361641.1
|
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr1_-_43638168 | 0.03 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr11_-_113577052 | 0.03 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr10_+_23384435 | 0.03 |
ENST00000376510.3
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr2_+_102615416 | 0.03 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chrX_+_11129388 | 0.03 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr14_+_35514323 | 0.03 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr18_-_51751132 | 0.03 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_+_12806141 | 0.03 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr6_+_149539767 | 0.03 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr1_+_65613340 | 0.03 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr18_+_21719018 | 0.02 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr12_+_57810198 | 0.02 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr7_-_105029812 | 0.02 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr14_+_20811722 | 0.02 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr17_+_33448593 | 0.02 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr7_-_76955563 | 0.02 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr4_-_122686261 | 0.02 |
ENST00000337677.5
|
TMEM155
|
transmembrane protein 155 |
| chr12_+_100594557 | 0.02 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr14_+_62164340 | 0.02 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr7_-_110174754 | 0.02 |
ENST00000435466.1
|
AC003088.1
|
AC003088.1 |
| chr15_+_57540230 | 0.02 |
ENST00000559703.1
|
TCF12
|
transcription factor 12 |
| chr5_+_158654712 | 0.02 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr7_+_36450169 | 0.02 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr14_+_74417192 | 0.02 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr19_+_21324827 | 0.02 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr1_-_200379104 | 0.02 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chrX_+_129040094 | 0.02 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr4_+_154622652 | 0.02 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr11_-_76381781 | 0.02 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr4_-_142134031 | 0.01 |
ENST00000420921.2
|
RNF150
|
ring finger protein 150 |
| chr9_-_2844058 | 0.01 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr20_-_48747662 | 0.01 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chrX_+_129040122 | 0.01 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr1_-_13331692 | 0.01 |
ENST00000353410.5
ENST00000376173.3 |
PRAMEF3
|
PRAME family member 3 |
| chr1_-_13698405 | 0.01 |
ENST00000540591.1
|
PRAMEF19
|
PRAME family member 19 |
| chr3_+_132316081 | 0.01 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr3_-_33686925 | 0.01 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_-_20801427 | 0.01 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr11_-_76381029 | 0.01 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr3_+_158362299 | 0.01 |
ENST00000478576.1
ENST00000264263.5 ENST00000464732.1 |
GFM1
|
G elongation factor, mitochondrial 1 |
| chr1_+_184020830 | 0.01 |
ENST00000533373.1
ENST00000423085.2 |
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr12_+_75874460 | 0.01 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr11_+_125496124 | 0.01 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr19_+_38880252 | 0.01 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr1_+_174769006 | 0.01 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_89641680 | 0.01 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr13_-_22178284 | 0.01 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr10_-_92681033 | 0.01 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_+_70876926 | 0.01 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr1_-_86848760 | 0.01 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr1_-_169455169 | 0.01 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr8_+_110551925 | 0.01 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr10_-_43892668 | 0.01 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr7_-_92848858 | 0.01 |
ENST00000440868.1
|
HEPACAM2
|
HEPACAM family member 2 |
| chr9_-_85882145 | 0.01 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr7_-_92848878 | 0.01 |
ENST00000341723.4
|
HEPACAM2
|
HEPACAM family member 2 |
| chr1_-_212004090 | 0.01 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr10_-_96829246 | 0.01 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_-_200379129 | 0.01 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr17_-_15519008 | 0.01 |
ENST00000261644.8
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_+_240255166 | 0.01 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr5_-_16916624 | 0.01 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr10_+_47894572 | 0.01 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr8_-_102181718 | 0.00 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr8_+_145133493 | 0.00 |
ENST00000316052.5
ENST00000525936.1 |
EXOSC4
|
exosome component 4 |
| chr16_-_11876408 | 0.00 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr4_+_156680153 | 0.00 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_+_202789394 | 0.00 |
ENST00000330493.5
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr11_+_125496400 | 0.00 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr14_+_62462541 | 0.00 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr20_-_14318248 | 0.00 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr2_+_87808725 | 0.00 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr2_+_11864458 | 0.00 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr17_-_15168624 | 0.00 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr14_-_74417096 | 0.00 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr2_+_171036635 | 0.00 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr2_+_24232918 | 0.00 |
ENST00000406420.3
ENST00000338315.4 |
MFSD2B
|
major facilitator superfamily domain containing 2B |
| chr1_+_32674675 | 0.00 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr22_-_16449805 | 0.00 |
ENST00000252835.4
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1 |
| chr6_+_123317116 | 0.00 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr12_-_110434183 | 0.00 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr4_-_74486347 | 0.00 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_13165457 | 0.00 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
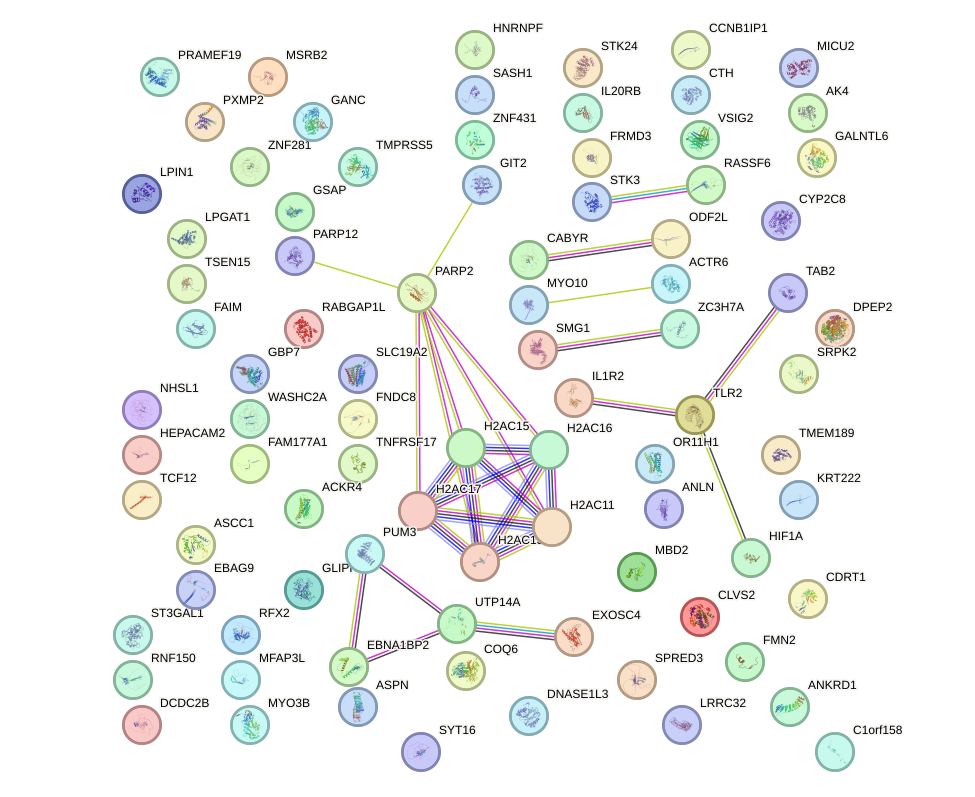
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |