Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for T
Z-value: 0.26
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSG00000164458.5 | T |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_99955042 | 0.09 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr3_+_195413160 | 0.08 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr2_+_102758210 | 0.07 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr2_+_102758751 | 0.06 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr15_+_101402041 | 0.06 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr17_+_26662679 | 0.05 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr20_-_44298878 | 0.05 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr6_+_131958436 | 0.05 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr7_-_81635106 | 0.05 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr14_+_85994943 | 0.04 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr1_+_19967014 | 0.04 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr12_+_54379569 | 0.04 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr19_+_6373715 | 0.04 |
ENST00000599849.1
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr17_+_47296865 | 0.04 |
ENST00000573347.1
|
ABI3
|
ABI family, member 3 |
| chr2_-_152118276 | 0.04 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr2_-_208030886 | 0.04 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_54378849 | 0.04 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr2_-_39664206 | 0.04 |
ENST00000484274.1
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr2_+_102758271 | 0.03 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr12_+_22778291 | 0.03 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr19_+_3721719 | 0.03 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr11_-_61196858 | 0.03 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr15_-_34502197 | 0.03 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr17_-_46657473 | 0.03 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr20_+_3190006 | 0.03 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr3_-_66551351 | 0.03 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr1_+_39670360 | 0.03 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr11_-_44972476 | 0.03 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr17_+_19281034 | 0.02 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr9_+_132044730 | 0.02 |
ENST00000455981.1
|
RP11-344B5.2
|
RP11-344B5.2 |
| chr5_-_137514333 | 0.02 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr2_+_108905095 | 0.02 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr5_-_138780159 | 0.02 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_-_61880248 | 0.02 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr10_-_76868931 | 0.02 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr6_-_33679452 | 0.02 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr22_+_35776828 | 0.02 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr7_-_28220354 | 0.02 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr1_-_85930246 | 0.02 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr2_-_216003127 | 0.02 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr21_+_30503282 | 0.02 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr2_+_162087577 | 0.02 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr3_+_142315294 | 0.02 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr16_-_88717423 | 0.02 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr19_-_1021113 | 0.02 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr10_+_94833642 | 0.02 |
ENST00000224356.4
ENST00000394139.1 |
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr21_-_27107283 | 0.02 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr2_+_192543694 | 0.02 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_14572070 | 0.02 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_102759199 | 0.02 |
ENST00000409288.1
ENST00000410023.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr17_-_33446820 | 0.02 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr4_-_4249924 | 0.02 |
ENST00000254742.2
ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128
|
transmembrane protein 128 |
| chr22_-_43567750 | 0.02 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr11_-_62995986 | 0.02 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr14_-_73997901 | 0.02 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr17_+_7533439 | 0.02 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr14_+_96722539 | 0.02 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr2_-_85636928 | 0.02 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr3_+_50654821 | 0.02 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_+_176972000 | 0.02 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr1_+_39456895 | 0.02 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr10_-_23003460 | 0.02 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr15_+_41913690 | 0.02 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr2_-_85895295 | 0.02 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr11_-_61197406 | 0.02 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_-_52002194 | 0.02 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr22_-_46644182 | 0.02 |
ENST00000404583.1
ENST00000404744.1 |
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr2_+_171571827 | 0.02 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr7_+_134576317 | 0.02 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr5_+_67576062 | 0.02 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_140005443 | 0.01 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr12_+_57610562 | 0.01 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr3_-_125094093 | 0.01 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr5_+_14664762 | 0.01 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr10_-_5060147 | 0.01 |
ENST00000604507.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr17_+_57970469 | 0.01 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr1_-_150980828 | 0.01 |
ENST00000361936.5
ENST00000361738.6 |
FAM63A
|
family with sequence similarity 63, member A |
| chr7_-_87849340 | 0.01 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chrX_-_106146547 | 0.01 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr4_+_26322409 | 0.01 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr9_+_36572851 | 0.01 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr2_+_33661382 | 0.01 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr15_-_34502278 | 0.01 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr2_+_234601512 | 0.01 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr7_+_110731062 | 0.01 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr3_+_167453493 | 0.01 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_+_27498289 | 0.01 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr11_-_236326 | 0.01 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr3_-_133614597 | 0.01 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr11_+_120081475 | 0.01 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr16_+_77233294 | 0.01 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr16_-_88717482 | 0.01 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr12_-_56710118 | 0.01 |
ENST00000273308.4
|
CNPY2
|
canopy FGF signaling regulator 2 |
| chr9_+_134378289 | 0.01 |
ENST00000423007.1
ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1
|
protein-O-mannosyltransferase 1 |
| chr4_+_128554081 | 0.01 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr17_-_61523622 | 0.01 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr12_+_19358228 | 0.01 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_-_229557 | 0.01 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr4_-_83934078 | 0.01 |
ENST00000505397.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr19_+_39647271 | 0.01 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr14_+_75746781 | 0.01 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_-_24126051 | 0.01 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr1_-_159832438 | 0.01 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr10_-_115423792 | 0.01 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr11_+_61197572 | 0.01 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr6_-_130031358 | 0.01 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr1_+_150980889 | 0.01 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr16_+_67313412 | 0.01 |
ENST00000379344.3
ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr12_-_90024360 | 0.01 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr22_-_32341336 | 0.01 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr17_-_17942473 | 0.01 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr11_+_61197508 | 0.01 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr7_-_81399355 | 0.01 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_120687335 | 0.01 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr11_-_61197187 | 0.01 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr9_-_117150303 | 0.01 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr19_-_17958771 | 0.01 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
| chr14_-_74025625 | 0.01 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr1_+_33219592 | 0.01 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr2_-_241737128 | 0.01 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr20_-_60942361 | 0.01 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr19_+_11546153 | 0.01 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr5_+_140557371 | 0.01 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr3_-_66551397 | 0.01 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr17_-_74497432 | 0.01 |
ENST00000590288.1
ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chrX_+_100353153 | 0.01 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr14_-_45603657 | 0.01 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr20_-_60942326 | 0.01 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr2_-_61245363 | 0.01 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr9_-_75567962 | 0.01 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr3_+_4535155 | 0.01 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr20_+_1115821 | 0.01 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr2_+_58134756 | 0.01 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr6_+_43149903 | 0.00 |
ENST00000252050.4
ENST00000354495.3 ENST00000372647.2 |
CUL9
|
cullin 9 |
| chr6_+_159290917 | 0.00 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr21_-_27107198 | 0.00 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr19_-_10514184 | 0.00 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr12_-_52867569 | 0.00 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr19_+_7895074 | 0.00 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr9_+_139553306 | 0.00 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr3_-_100712292 | 0.00 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr5_+_139554227 | 0.00 |
ENST00000261811.4
|
CYSTM1
|
cysteine-rich transmembrane module containing 1 |
| chr1_+_17516275 | 0.00 |
ENST00000412427.1
|
RP11-380J14.1
|
RP11-380J14.1 |
| chr1_+_35225339 | 0.00 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr9_+_79074068 | 0.00 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr12_-_56727676 | 0.00 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr16_+_66429358 | 0.00 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr14_+_95078714 | 0.00 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr11_+_111749650 | 0.00 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr12_-_6961050 | 0.00 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr6_-_97731019 | 0.00 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr16_+_67596310 | 0.00 |
ENST00000264010.4
ENST00000401394.1 |
CTCF
|
CCCTC-binding factor (zinc finger protein) |
| chr7_-_752577 | 0.00 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr10_-_79789291 | 0.00 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr14_-_69619689 | 0.00 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr1_-_45452240 | 0.00 |
ENST00000372183.3
ENST00000372182.4 ENST00000360403.2 |
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa |
| chr5_+_67576109 | 0.00 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_54378923 | 0.00 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr11_-_66112555 | 0.00 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr2_+_27498331 | 0.00 |
ENST00000402462.1
ENST00000404433.1 ENST00000406962.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr3_-_100712352 | 0.00 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_+_57741957 | 0.00 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr7_-_994302 | 0.00 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr11_+_111412271 | 0.00 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr15_+_78832747 | 0.00 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr2_-_208030647 | 0.00 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_115124257 | 0.00 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr4_-_151936865 | 0.00 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr11_-_61196752 | 0.00 |
ENST00000448745.1
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr8_-_114449112 | 0.00 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr5_+_85913721 | 0.00 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr19_+_58258164 | 0.00 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr17_+_17942684 | 0.00 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr2_+_189839046 | 0.00 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr1_+_78245303 | 0.00 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr2_+_166152283 | 0.00 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr8_+_7801144 | 0.00 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr19_-_18366182 | 0.00 |
ENST00000355502.3
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr21_-_27107344 | 0.00 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_+_39670423 | 0.00 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr14_-_69619823 | 0.00 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr2_-_27341966 | 0.00 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr20_+_32951070 | 0.00 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr14_-_69619291 | 0.00 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr1_+_114472481 | 0.00 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_+_26158343 | 0.00 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr10_-_5060201 | 0.00 |
ENST00000407674.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
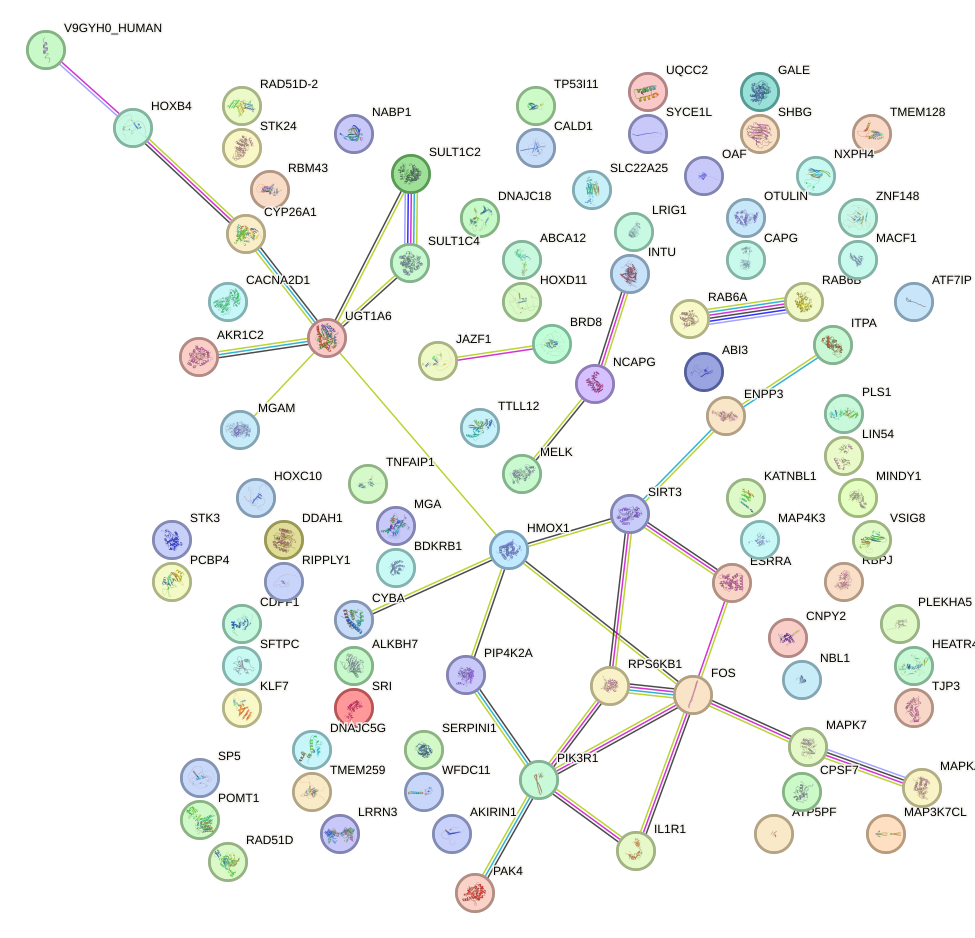
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |