Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for TCF7L1
Z-value: 0.50
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | transcription factor 7 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L1 | hg19_v2_chr2_+_85360499_85360598 | -0.31 | 6.9e-01 | Click! |
Activity profile of TCF7L1 motif
Sorted Z-values of TCF7L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_102200948 | 0.46 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_233734994 | 0.44 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr19_+_51152702 | 0.43 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr17_-_7297833 | 0.37 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr16_+_2820912 | 0.35 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr17_+_62223320 | 0.33 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr7_-_105926058 | 0.33 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr12_-_108154705 | 0.33 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr1_+_16085263 | 0.31 |
ENST00000483633.2
ENST00000502739.1 ENST00000431771.2 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_+_79679299 | 0.31 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr14_-_23791484 | 0.30 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr15_-_90294523 | 0.29 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr14_-_20922960 | 0.29 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr17_+_72428266 | 0.28 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr6_-_32145861 | 0.25 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr19_-_59030921 | 0.25 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr11_+_65339820 | 0.24 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr12_-_49453557 | 0.23 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr11_-_65381643 | 0.22 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr17_+_72428218 | 0.22 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr19_-_59031118 | 0.22 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr13_-_30160925 | 0.21 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_-_175964366 | 0.21 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr15_+_81293254 | 0.20 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr19_-_7990991 | 0.19 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr5_+_175288631 | 0.19 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_1334685 | 0.18 |
ENST00000400809.3
ENST00000408918.4 |
CCNL2
|
cyclin L2 |
| chr9_-_27005686 | 0.17 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr8_+_17104401 | 0.17 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr12_-_130529501 | 0.17 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr18_+_3450161 | 0.17 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_16085244 | 0.16 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr14_-_23451845 | 0.16 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr11_-_117747327 | 0.16 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr2_-_97534312 | 0.16 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr7_+_150076406 | 0.16 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr14_-_23451467 | 0.15 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr12_+_28605426 | 0.15 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr17_-_7297519 | 0.15 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr16_-_73082274 | 0.15 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr3_-_148939598 | 0.15 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_-_4852915 | 0.15 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr1_-_43424500 | 0.15 |
ENST00000415851.2
ENST00000426263.3 ENST00000372500.3 |
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr19_+_41257084 | 0.14 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr16_+_30406423 | 0.14 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr6_-_33548006 | 0.14 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr14_-_65409502 | 0.14 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr11_+_65999265 | 0.14 |
ENST00000528935.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_-_145047688 | 0.14 |
ENST00000356346.3
|
PLEC
|
plectin |
| chr17_-_46035187 | 0.13 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chrX_+_54835493 | 0.13 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr10_-_104192405 | 0.13 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr12_+_1738363 | 0.12 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr19_-_3061397 | 0.12 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr9_+_139221880 | 0.12 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr22_-_31688431 | 0.12 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_-_62474803 | 0.12 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_+_53491220 | 0.11 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr14_+_105941118 | 0.11 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr12_-_49449107 | 0.11 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_80275953 | 0.11 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr18_-_56985776 | 0.11 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr1_-_32801825 | 0.11 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr10_+_54074033 | 0.11 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr15_+_90744745 | 0.11 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr20_+_36149602 | 0.11 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr11_-_46615498 | 0.10 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr20_+_55966444 | 0.10 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr19_-_40791302 | 0.10 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr10_-_101380121 | 0.10 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr6_-_33547975 | 0.10 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_-_11450249 | 0.10 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr1_-_219615984 | 0.10 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr12_-_49582978 | 0.09 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr19_+_51153045 | 0.09 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr12_+_54694979 | 0.09 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr6_-_107436473 | 0.09 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr2_+_47168630 | 0.09 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr19_+_1249869 | 0.09 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr9_+_109685630 | 0.09 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr17_+_80416050 | 0.09 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr1_+_36348790 | 0.09 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr5_-_58335281 | 0.09 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr15_-_64995399 | 0.09 |
ENST00000559753.1
ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr12_-_49582837 | 0.08 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_6479963 | 0.08 |
ENST00000377836.4
ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2
|
hes family bHLH transcription factor 2 |
| chr17_-_39093672 | 0.08 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr3_+_152879985 | 0.08 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr20_+_44509857 | 0.08 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr2_+_196440692 | 0.08 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr22_-_31688381 | 0.07 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr10_+_71561630 | 0.07 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_-_140477910 | 0.07 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_200992827 | 0.07 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr1_+_8378140 | 0.07 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr7_+_73245193 | 0.07 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr2_+_47168313 | 0.07 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr8_-_11660077 | 0.07 |
ENST00000533405.1
|
RP11-297N6.4
|
Uncharacterized protein |
| chrX_-_128788914 | 0.07 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr4_-_120243545 | 0.07 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr7_+_73703728 | 0.07 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr1_+_87794150 | 0.07 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr16_+_57673348 | 0.06 |
ENST00000567915.1
ENST00000564103.1 ENST00000562467.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_+_55354822 | 0.06 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr7_+_128828713 | 0.06 |
ENST00000249373.3
|
SMO
|
smoothened, frizzled family receptor |
| chr16_+_30406721 | 0.06 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chrX_-_153599578 | 0.06 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr12_+_9066472 | 0.05 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr15_+_65134088 | 0.05 |
ENST00000323544.4
ENST00000437723.1 |
PLEKHO2
AC069368.3
|
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr12_+_109915179 | 0.05 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr20_-_656437 | 0.05 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr15_+_75074410 | 0.05 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr1_+_59762642 | 0.05 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr17_+_80416482 | 0.05 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr1_-_231005310 | 0.05 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr18_+_3449821 | 0.05 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_106301405 | 0.05 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr11_-_87908600 | 0.05 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chrX_+_152760397 | 0.05 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chr15_+_80696666 | 0.05 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr7_+_139528952 | 0.05 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr5_-_157002775 | 0.05 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr5_-_138725560 | 0.04 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr17_+_42923686 | 0.04 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr2_+_27071292 | 0.04 |
ENST00000431402.1
ENST00000434719.1 |
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr11_-_75921780 | 0.04 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr12_-_58146128 | 0.04 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr2_+_179184955 | 0.04 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr19_+_41256764 | 0.04 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_+_22605304 | 0.04 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr8_-_33424636 | 0.04 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr10_+_114710425 | 0.04 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_8274204 | 0.04 |
ENST00000561053.1
ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4
|
ceramide synthase 4 |
| chr1_-_109935819 | 0.04 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr13_+_39612485 | 0.04 |
ENST00000379599.2
|
NHLRC3
|
NHL repeat containing 3 |
| chr22_+_41347363 | 0.04 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr17_+_80416609 | 0.04 |
ENST00000577410.1
|
NARF
|
nuclear prelamin A recognition factor |
| chr12_+_56324933 | 0.04 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr15_+_75074915 | 0.04 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr8_-_124553437 | 0.03 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chrX_-_110655391 | 0.03 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr6_+_134274354 | 0.03 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr10_+_11206925 | 0.03 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_+_35851570 | 0.03 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr6_+_143929307 | 0.03 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr2_-_165697717 | 0.03 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr3_-_149375783 | 0.03 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr15_+_43885799 | 0.03 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_-_61819229 | 0.03 |
ENST00000447001.3
ENST00000392950.4 |
STRADA
|
STE20-related kinase adaptor alpha |
| chr7_-_27219849 | 0.03 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr13_+_24144796 | 0.03 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_-_166060552 | 0.03 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr6_+_35704855 | 0.03 |
ENST00000288065.2
ENST00000373866.3 |
ARMC12
|
armadillo repeat containing 12 |
| chr19_-_40791211 | 0.03 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_+_70026795 | 0.03 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr1_+_223101757 | 0.03 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr13_+_24144509 | 0.03 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chrX_+_106163626 | 0.03 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr13_+_30002741 | 0.03 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr19_+_8274185 | 0.03 |
ENST00000558268.1
ENST00000558331.1 |
CERS4
|
ceramide synthase 4 |
| chr14_+_75746664 | 0.03 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr6_+_12008986 | 0.03 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr4_-_69215699 | 0.03 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr14_-_73493825 | 0.03 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr1_-_20126365 | 0.03 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr12_-_49582593 | 0.03 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr8_+_21777243 | 0.03 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr6_+_119215308 | 0.03 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr4_-_139163491 | 0.02 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_+_180199393 | 0.02 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr4_+_79567362 | 0.02 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr1_+_154244987 | 0.02 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chrX_-_110655306 | 0.02 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr9_+_124088860 | 0.02 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr2_-_166060571 | 0.02 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr10_+_71561704 | 0.02 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr1_-_94079648 | 0.02 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr9_-_74525847 | 0.02 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr11_+_69455855 | 0.02 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr10_+_71561649 | 0.02 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chrX_+_69488174 | 0.02 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr13_+_30002846 | 0.02 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr12_+_80603233 | 0.02 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr10_+_71562180 | 0.02 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_-_18717627 | 0.02 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr11_+_77184416 | 0.02 |
ENST00000598970.1
|
DKFZP434E1119
|
DKFZP434E1119 |
| chr8_-_128231299 | 0.02 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr10_+_22605374 | 0.02 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr11_+_36317830 | 0.02 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr8_-_29939933 | 0.02 |
ENST00000522794.1
|
TMEM66
|
transmembrane protein 66 |
| chr7_-_27213893 | 0.02 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr6_+_108881012 | 0.02 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr14_+_39644425 | 0.01 |
ENST00000556530.1
|
PNN
|
pinin, desmosome associated protein |
| chr2_+_48796120 | 0.01 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr3_-_47823298 | 0.01 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr12_-_109915098 | 0.01 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr5_+_102201430 | 0.01 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_191273052 | 0.01 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
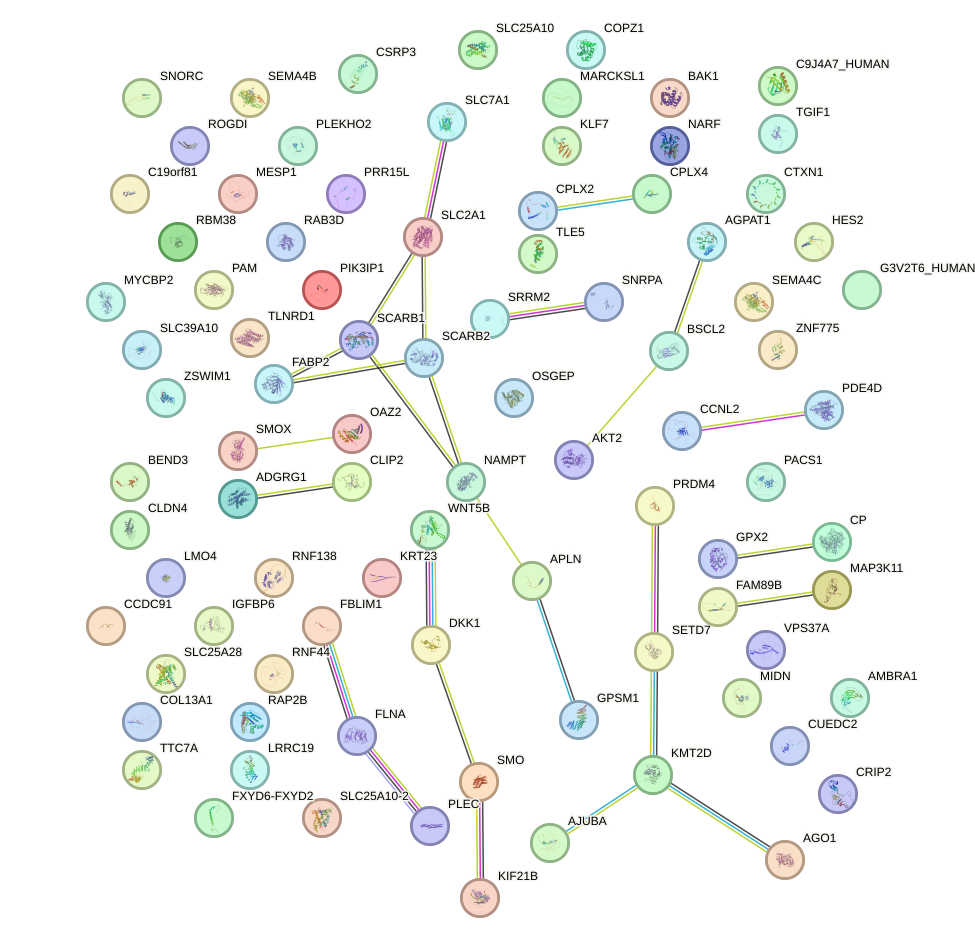
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.1 | 0.3 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.2 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |