Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
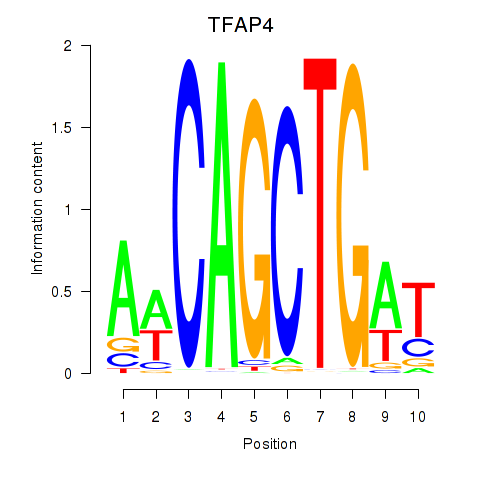
Results for TFAP4_MSC
Z-value: 0.51
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.7 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.8 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | 0.86 | 1.4e-01 | Click! |
| MSC | hg19_v2_chr8_-_72756667_72756736 | -0.58 | 4.2e-01 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_68845417 | 0.55 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr4_+_108815402 | 0.31 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr12_-_123752624 | 0.27 |
ENST00000542174.1
ENST00000535796.1 |
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr17_+_36508826 | 0.23 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr3_-_52869205 | 0.22 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr8_-_71519889 | 0.16 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr17_-_15469590 | 0.16 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr1_-_150669604 | 0.16 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr3_+_69134124 | 0.16 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr2_+_136343904 | 0.15 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr4_-_46391367 | 0.15 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr13_+_41635617 | 0.14 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr2_+_233404429 | 0.14 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr6_+_44184653 | 0.13 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr1_+_212738676 | 0.13 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr4_-_52904425 | 0.13 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chrX_-_108868390 | 0.12 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr5_+_176692466 | 0.12 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr2_-_17981462 | 0.12 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr8_-_74791051 | 0.12 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr4_+_37892682 | 0.12 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr19_-_39735646 | 0.12 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr5_+_66124590 | 0.11 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_19804973 | 0.11 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr9_+_117350009 | 0.11 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr11_+_61722629 | 0.11 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chr18_-_19180681 | 0.11 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr19_-_13068012 | 0.11 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr1_+_46379254 | 0.11 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr8_+_81397846 | 0.11 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr12_+_652294 | 0.10 |
ENST00000322843.3
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr15_+_83776137 | 0.10 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr2_+_33701707 | 0.10 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_14075865 | 0.10 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr3_+_159570722 | 0.10 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr8_-_99837856 | 0.10 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr14_-_23284675 | 0.09 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_-_40349106 | 0.09 |
ENST00000545233.1
ENST00000537440.1 ENST00000537223.1 ENST00000541099.1 ENST00000441669.2 ENST00000544981.1 ENST00000316891.5 ENST00000372818.1 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chr11_-_85430204 | 0.09 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr9_-_16705069 | 0.09 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr5_+_49962772 | 0.09 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr16_+_23847339 | 0.09 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr2_+_136343820 | 0.09 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr15_+_49715293 | 0.09 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr2_-_176866978 | 0.09 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr9_+_125795788 | 0.09 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr12_-_57644952 | 0.09 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr2_+_233562015 | 0.08 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr1_+_78354175 | 0.08 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr7_-_120497178 | 0.08 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr9_+_6758024 | 0.08 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr11_-_85430356 | 0.08 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr15_-_67546963 | 0.08 |
ENST00000561452.1
ENST00000261880.5 |
AAGAB
|
alpha- and gamma-adaptin binding protein |
| chr15_-_75748115 | 0.08 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr15_+_93447675 | 0.08 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chrX_+_15756382 | 0.08 |
ENST00000318636.3
|
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr10_+_94594351 | 0.08 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr16_-_4323015 | 0.08 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr5_+_72509751 | 0.08 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr4_-_186125077 | 0.07 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr17_+_73028676 | 0.07 |
ENST00000581589.1
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr5_+_102201509 | 0.07 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_174844645 | 0.07 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr15_-_52263937 | 0.07 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr21_-_10990830 | 0.07 |
ENST00000361285.4
ENST00000342420.5 ENST00000328758.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr2_+_30370382 | 0.07 |
ENST00000402708.1
|
YPEL5
|
yippee-like 5 (Drosophila) |
| chr17_+_41003166 | 0.07 |
ENST00000308423.2
|
AOC3
|
amine oxidase, copper containing 3 |
| chr21_+_37692481 | 0.07 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr3_+_69134080 | 0.07 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr19_+_35521699 | 0.07 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr15_-_75748143 | 0.07 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr8_+_38758845 | 0.07 |
ENST00000519640.1
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr3_+_32148106 | 0.07 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr11_+_77774897 | 0.07 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr2_-_42180940 | 0.07 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr12_-_118628315 | 0.07 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr4_-_74864386 | 0.07 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr5_-_16936340 | 0.07 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr1_+_78354297 | 0.07 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_47603109 | 0.07 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr19_+_35521616 | 0.07 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr2_+_136343943 | 0.07 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr3_+_167453026 | 0.07 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr3_-_81792780 | 0.07 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr6_-_108582168 | 0.07 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr5_-_58571935 | 0.07 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_-_49118067 | 0.07 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83, member E |
| chr2_-_61245363 | 0.06 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr1_+_153600869 | 0.06 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr17_+_44679808 | 0.06 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr20_-_3687775 | 0.06 |
ENST00000344754.4
ENST00000202578.4 |
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr20_+_33292507 | 0.06 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_33336414 | 0.06 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr6_+_155537771 | 0.06 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_+_172422026 | 0.06 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr8_-_66701319 | 0.06 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr14_-_71107921 | 0.06 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr7_-_30029367 | 0.06 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr7_-_30029574 | 0.06 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr1_-_206945830 | 0.06 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr2_+_192110199 | 0.06 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr14_+_62162258 | 0.06 |
ENST00000337138.4
ENST00000394997.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr17_+_12693001 | 0.06 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr4_+_78079570 | 0.06 |
ENST00000509972.1
|
CCNG2
|
cyclin G2 |
| chr15_-_42749711 | 0.06 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr15_+_81299370 | 0.06 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr8_-_30670053 | 0.06 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr15_+_65337708 | 0.06 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr4_-_185655212 | 0.06 |
ENST00000541971.1
|
MLF1IP
|
centromere protein U |
| chr22_+_38071615 | 0.06 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr10_+_13142225 | 0.06 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr9_+_6757634 | 0.06 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr2_-_202645835 | 0.06 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr17_-_42345487 | 0.05 |
ENST00000262418.6
|
SLC4A1
|
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr2_+_200472779 | 0.05 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chrX_+_70521584 | 0.05 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr1_-_154164534 | 0.05 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr12_-_70093235 | 0.05 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr1_-_150849047 | 0.05 |
ENST00000354396.2
ENST00000505755.1 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr8_+_71581565 | 0.05 |
ENST00000408926.3
ENST00000520030.1 |
XKR9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr10_-_97321112 | 0.05 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr19_-_2151523 | 0.05 |
ENST00000350812.6
ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr7_+_73507409 | 0.05 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr10_-_29923893 | 0.05 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_15272271 | 0.05 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr2_+_135011731 | 0.05 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr14_-_53019211 | 0.05 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr5_+_54455946 | 0.05 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr3_+_32147997 | 0.05 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr15_+_57998923 | 0.05 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr7_-_108166505 | 0.05 |
ENST00000426128.2
ENST00000427008.1 ENST00000388728.5 ENST00000257694.8 ENST00000422087.1 ENST00000453144.1 ENST00000436062.1 |
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr11_+_7597639 | 0.05 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr11_-_65629497 | 0.05 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr17_+_58018269 | 0.05 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr17_+_76037081 | 0.05 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr3_-_65583561 | 0.05 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr19_+_11651942 | 0.05 |
ENST00000587087.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr17_+_7462103 | 0.05 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_101437487 | 0.05 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr11_-_117747434 | 0.05 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_56221330 | 0.05 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr2_+_85822857 | 0.05 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr4_-_89619386 | 0.05 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr9_-_21305312 | 0.05 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr12_+_133033703 | 0.05 |
ENST00000542627.1
|
RP11-503G7.1
|
RP11-503G7.1 |
| chr12_+_78224667 | 0.05 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr19_+_32836499 | 0.05 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr5_+_102201722 | 0.05 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_+_8316442 | 0.05 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr5_+_145583107 | 0.05 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr19_+_35521572 | 0.05 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr3_+_99833755 | 0.05 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr22_-_36924944 | 0.05 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr10_-_102090243 | 0.05 |
ENST00000338519.3
ENST00000353274.3 ENST00000318222.3 |
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr17_+_7462031 | 0.05 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr15_+_67547113 | 0.05 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr14_+_90863327 | 0.05 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr6_-_24911195 | 0.05 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr5_-_141257954 | 0.05 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr15_+_67547163 | 0.04 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr8_-_71581377 | 0.04 |
ENST00000276590.4
ENST00000522447.1 |
LACTB2
|
lactamase, beta 2 |
| chr19_+_7587555 | 0.04 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr3_-_33700933 | 0.04 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_102201687 | 0.04 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_+_123430948 | 0.04 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chrX_+_69509927 | 0.04 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr9_+_35792151 | 0.04 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr6_-_19804877 | 0.04 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr10_-_92681033 | 0.04 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_-_150669500 | 0.04 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_-_85462623 | 0.04 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr17_+_33570055 | 0.04 |
ENST00000299977.4
ENST00000542451.1 ENST00000592325.1 |
SLFN5
|
schlafen family member 5 |
| chr17_-_57232525 | 0.04 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_+_56473628 | 0.04 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_+_7506713 | 0.04 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr17_+_67957878 | 0.04 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr22_+_19705928 | 0.04 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr1_-_150849174 | 0.04 |
ENST00000515192.1
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr18_-_55470320 | 0.04 |
ENST00000536015.1
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr3_+_182511266 | 0.04 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr12_+_57828521 | 0.04 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr10_+_120863587 | 0.04 |
ENST00000535029.1
ENST00000361432.2 ENST00000544016.1 |
FAM45A
|
family with sequence similarity 45, member A |
| chr6_+_17281573 | 0.04 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr4_-_80994210 | 0.04 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr12_-_3862245 | 0.04 |
ENST00000252322.1
ENST00000440314.2 |
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr11_+_71259466 | 0.04 |
ENST00000528743.2
|
KRTAP5-9
|
keratin associated protein 5-9 |
| chr4_-_177198663 | 0.04 |
ENST00000505299.1
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr5_+_145583156 | 0.04 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr11_-_64013288 | 0.04 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_-_173173171 | 0.04 |
ENST00000517733.1
ENST00000518902.1 |
CTB-43E15.3
|
CTB-43E15.3 |
| chr16_+_85204882 | 0.04 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr6_-_76203345 | 0.04 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_+_7341586 | 0.04 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr1_+_109756523 | 0.04 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr1_+_178482262 | 0.04 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr10_-_111683308 | 0.04 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr2_-_183903133 | 0.04 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr11_+_71498552 | 0.04 |
ENST00000346333.6
ENST00000359244.4 ENST00000426628.2 |
FAM86C1
|
family with sequence similarity 86, member C1 |
| chr12_-_4647606 | 0.04 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr21_-_27543425 | 0.04 |
ENST00000448388.2
|
APP
|
amyloid beta (A4) precursor protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.2 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |