Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
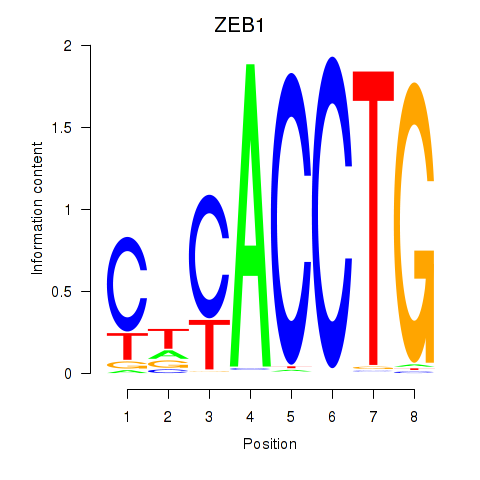
Results for ZEB1
Z-value: 0.68
Transcription factors associated with ZEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZEB1
|
ENSG00000148516.17 | zinc finger E-box binding homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZEB1 | hg19_v2_chr10_+_31608054_31608156 | -0.97 | 3.2e-02 | Click! |
Activity profile of ZEB1 motif
Sorted Z-values of ZEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_6464243 | 0.70 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr16_+_771663 | 0.46 |
ENST00000568916.1
|
FAM173A
|
family with sequence similarity 173, member A |
| chr19_+_6464502 | 0.43 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr9_-_131940526 | 0.40 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr8_+_21906433 | 0.40 |
ENST00000522148.1
|
DMTN
|
dematin actin binding protein |
| chr22_-_21984282 | 0.39 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr11_+_394145 | 0.38 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr8_+_22429205 | 0.38 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr11_-_417308 | 0.36 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr2_+_220306238 | 0.34 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr16_+_691792 | 0.33 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr8_-_144815966 | 0.33 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chr12_+_7072354 | 0.33 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr12_-_25150373 | 0.32 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr16_-_67190152 | 0.31 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr22_+_43547937 | 0.30 |
ENST00000329563.4
|
TSPO
|
translocator protein (18kDa) |
| chr19_-_6767431 | 0.30 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr17_+_72428266 | 0.29 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr10_+_99258625 | 0.29 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr19_-_40324767 | 0.29 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr7_-_73184588 | 0.28 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr17_+_3572087 | 0.28 |
ENST00000248378.5
ENST00000397133.2 |
EMC6
|
ER membrane protein complex subunit 6 |
| chr11_-_417388 | 0.28 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr11_-_560703 | 0.28 |
ENST00000441853.1
ENST00000329451.3 |
C11orf35
|
chromosome 11 open reading frame 35 |
| chr16_+_3070313 | 0.27 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr1_-_27226928 | 0.27 |
ENST00000361720.5
|
GPATCH3
|
G patch domain containing 3 |
| chr19_-_6767516 | 0.27 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr16_+_3068393 | 0.26 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr19_-_10697895 | 0.26 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr1_+_16085263 | 0.26 |
ENST00000483633.2
ENST00000502739.1 ENST00000431771.2 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_-_32827682 | 0.26 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr17_+_74732889 | 0.24 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr16_-_89752965 | 0.24 |
ENST00000567544.1
|
RP11-368I7.4
|
Uncharacterized protein |
| chr17_+_73521763 | 0.24 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr4_+_76649753 | 0.24 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr10_-_81932771 | 0.24 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr15_+_89182156 | 0.24 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr17_+_72428218 | 0.23 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr19_+_35739897 | 0.23 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr17_-_39942322 | 0.23 |
ENST00000449889.1
ENST00000465293.1 |
JUP
|
junction plakoglobin |
| chr19_-_55895966 | 0.23 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr17_+_7123207 | 0.22 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr6_+_53659877 | 0.22 |
ENST00000370882.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr19_+_35739631 | 0.22 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_233734994 | 0.22 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr5_+_176873789 | 0.22 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr19_-_55791058 | 0.22 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr16_-_1821496 | 0.21 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_-_172662303 | 0.21 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr16_-_2260834 | 0.21 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr17_-_61512545 | 0.21 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr8_-_145754428 | 0.20 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr1_+_31885963 | 0.20 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr8_+_145438870 | 0.20 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr7_+_73106926 | 0.20 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr3_-_107941209 | 0.20 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr16_+_3070356 | 0.20 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr19_+_36036477 | 0.20 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr19_-_55791431 | 0.20 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_-_27230035 | 0.20 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr5_+_149865838 | 0.20 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr14_-_69262947 | 0.19 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_18266818 | 0.19 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr14_-_61124977 | 0.19 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr9_+_130922537 | 0.19 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr11_+_62379194 | 0.19 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr11_+_394196 | 0.19 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr16_-_3030407 | 0.19 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr1_+_38273419 | 0.19 |
ENST00000468084.1
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr8_+_22844913 | 0.19 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr7_-_139168402 | 0.19 |
ENST00000393039.2
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr17_-_66097610 | 0.19 |
ENST00000584047.1
ENST00000579629.1 |
AC145343.2
|
AC145343.2 |
| chr19_-_40324255 | 0.19 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr1_+_200863949 | 0.18 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr15_-_83224682 | 0.18 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr9_-_117160738 | 0.18 |
ENST00000448674.1
|
RP11-9M16.2
|
RP11-9M16.2 |
| chr19_-_14016877 | 0.18 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr6_+_44095263 | 0.18 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr19_-_50316423 | 0.18 |
ENST00000528094.1
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr1_+_154975110 | 0.18 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_-_776431 | 0.18 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr19_+_51152702 | 0.18 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr9_-_131486367 | 0.18 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr17_+_2264983 | 0.18 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr19_+_3539152 | 0.18 |
ENST00000329493.5
|
C19orf71
|
chromosome 19 open reading frame 71 |
| chr5_-_133747551 | 0.18 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr3_-_49055991 | 0.18 |
ENST00000441576.2
ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr8_-_21966893 | 0.18 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr7_-_97881429 | 0.17 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr2_+_130939235 | 0.17 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr11_+_560956 | 0.17 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr5_-_140053152 | 0.17 |
ENST00000542735.1
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr19_-_7990991 | 0.17 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr17_+_42422629 | 0.17 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr12_-_53625958 | 0.17 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr8_+_22436248 | 0.17 |
ENST00000308354.7
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr16_+_616995 | 0.17 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chr11_+_66624527 | 0.17 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_+_1823208 | 0.17 |
ENST00000568449.1
ENST00000307394.7 |
EME2
|
essential meiotic structure-specific endonuclease subunit 2 |
| chr19_-_8408139 | 0.17 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr5_+_176873446 | 0.17 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr6_-_31612808 | 0.17 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr3_-_121740969 | 0.17 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr16_-_67194201 | 0.17 |
ENST00000345057.4
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr8_+_22428457 | 0.17 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_87040259 | 0.17 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr17_+_21188012 | 0.17 |
ENST00000529517.1
|
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr14_-_69262789 | 0.17 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_+_61100676 | 0.17 |
ENST00000394900.3
ENST00000532173.2 |
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae) |
| chr8_+_143808605 | 0.16 |
ENST00000336138.3
|
THEM6
|
thioesterase superfamily member 6 |
| chr16_+_1832902 | 0.16 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr19_-_19314162 | 0.16 |
ENST00000420605.3
ENST00000544883.1 ENST00000538165.2 ENST00000331552.7 |
NR2C2AP
|
nuclear receptor 2C2-associated protein |
| chr17_+_80014359 | 0.16 |
ENST00000578168.1
|
GPS1
|
G protein pathway suppressor 1 |
| chr15_+_76030311 | 0.16 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr1_-_229569834 | 0.16 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr19_+_14544099 | 0.16 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr11_+_66610883 | 0.16 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr14_-_69262916 | 0.16 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_-_10420459 | 0.16 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr16_-_3030283 | 0.16 |
ENST00000572619.1
ENST00000574415.1 ENST00000440027.2 ENST00000572059.1 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr19_+_38755237 | 0.16 |
ENST00000587516.1
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr16_-_30107491 | 0.16 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr17_-_39942940 | 0.16 |
ENST00000310706.5
ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP
|
junction plakoglobin |
| chr1_+_38273818 | 0.16 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr7_+_2687173 | 0.16 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr17_-_79519403 | 0.16 |
ENST00000327787.8
ENST00000537152.1 |
C17orf70
|
chromosome 17 open reading frame 70 |
| chr14_+_38065052 | 0.16 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr22_-_50970506 | 0.16 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_+_38465441 | 0.15 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr16_+_2820912 | 0.15 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr11_-_67236691 | 0.15 |
ENST00000544903.1
ENST00000308022.2 ENST00000393877.3 ENST00000452789.2 |
TMEM134
|
transmembrane protein 134 |
| chr19_+_1905365 | 0.15 |
ENST00000329478.2
ENST00000602400.1 ENST00000409472.1 |
ADAT3
SCAMP4
|
adenosine deaminase, tRNA-specific 3 secretory carrier membrane protein 4 |
| chrX_-_153718988 | 0.15 |
ENST00000263512.4
ENST00000393587.4 ENST00000453912.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr14_-_24732738 | 0.15 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr19_-_47735918 | 0.15 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr8_+_22436635 | 0.15 |
ENST00000452226.1
ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_16085244 | 0.15 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_-_167906020 | 0.15 |
ENST00000458574.1
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr20_-_62199427 | 0.15 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr19_+_50016411 | 0.15 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr18_+_21594384 | 0.15 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr9_-_131790550 | 0.15 |
ENST00000372554.4
ENST00000372564.3 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr17_+_80193644 | 0.15 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr16_+_57653989 | 0.15 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr14_+_75745477 | 0.15 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr17_-_4607335 | 0.15 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr12_-_123450986 | 0.15 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr8_+_144816303 | 0.15 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr16_+_89753070 | 0.14 |
ENST00000353379.7
ENST00000505473.1 ENST00000564192.1 |
CDK10
|
cyclin-dependent kinase 10 |
| chr19_+_36236491 | 0.14 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr17_-_41623716 | 0.14 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr16_+_3019309 | 0.14 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chrX_+_47050798 | 0.14 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr8_-_145752390 | 0.14 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr17_+_42422662 | 0.14 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr12_-_49365501 | 0.14 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr22_+_45098067 | 0.14 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr19_-_17366257 | 0.14 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr19_-_14316980 | 0.14 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr15_+_75640068 | 0.14 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr8_-_145742862 | 0.14 |
ENST00000524998.1
|
RECQL4
|
RecQ protein-like 4 |
| chr10_+_71389983 | 0.14 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chr19_+_36235964 | 0.14 |
ENST00000587708.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr14_-_65409502 | 0.14 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr17_-_31204124 | 0.14 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr11_-_47600549 | 0.14 |
ENST00000430070.2
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr1_+_24120143 | 0.14 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr11_-_18270182 | 0.14 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr19_+_50919056 | 0.13 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr1_-_41328018 | 0.13 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr2_+_90198535 | 0.13 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr8_+_145734433 | 0.13 |
ENST00000301327.4
|
MFSD3
|
major facilitator superfamily domain containing 3 |
| chr14_-_80677815 | 0.13 |
ENST00000557125.1
ENST00000555750.1 |
DIO2
|
deiodinase, iodothyronine, type II |
| chr6_-_42016385 | 0.13 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr7_+_127527965 | 0.13 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr16_-_31106211 | 0.13 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr22_+_29469012 | 0.13 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr9_-_139940608 | 0.13 |
ENST00000371601.4
|
NPDC1
|
neural proliferation, differentiation and control, 1 |
| chr11_-_65314905 | 0.13 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr17_-_7145106 | 0.13 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr16_+_3019246 | 0.13 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr9_+_130159504 | 0.13 |
ENST00000373352.1
ENST00000373360.3 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr19_+_36239576 | 0.13 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr2_+_241526126 | 0.13 |
ENST00000391984.2
ENST00000391982.2 ENST00000404753.3 ENST00000270364.7 ENST00000352879.4 ENST00000354082.4 |
CAPN10
|
calpain 10 |
| chr16_+_57653854 | 0.13 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_-_88878305 | 0.13 |
ENST00000569616.1
ENST00000563655.1 ENST00000567713.1 ENST00000426324.2 ENST00000378364.3 |
APRT
|
adenine phosphoribosyltransferase |
| chr15_+_41245160 | 0.13 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr2_-_220408260 | 0.13 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr17_-_40346477 | 0.13 |
ENST00000593209.1
ENST00000587427.1 ENST00000588352.1 ENST00000414034.3 ENST00000590249.1 |
GHDC
|
GH3 domain containing |
| chr22_+_29702996 | 0.13 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr19_+_13228917 | 0.13 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr10_-_99258135 | 0.13 |
ENST00000327238.10
ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr11_-_61129723 | 0.13 |
ENST00000537680.1
ENST00000426130.2 ENST00000294072.4 |
CYB561A3
|
cytochrome b561 family, member A3 |
| chr10_+_104221137 | 0.13 |
ENST00000366277.2
ENST00000238936.4 ENST00000369931.3 |
TMEM180
|
transmembrane protein 180 |
| chr15_-_75660919 | 0.13 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr11_+_45825616 | 0.13 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr19_+_36236514 | 0.13 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr12_+_57916584 | 0.13 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_+_34175398 | 0.13 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr16_-_28223229 | 0.13 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.4 | GO:0060929 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.1 | 0.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.4 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.7 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 0.1 | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.1 | 0.2 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of fertilization(GO:0060467) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.3 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.5 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.0 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0060482 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.0 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:1902403 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.0 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.0 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.0 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.0 | 0.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.0 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.0 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) divalent metal ion export(GO:0070839) arginine import(GO:0090467) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.0 | GO:0002415 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.0 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.0 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0052419 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.0 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 0.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.0 | GO:0010820 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0019050 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.0 | 0.0 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.4 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.4 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.0 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.0 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.0 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |