Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AGUGCUU
Z-value: 0.61
miRNA associated with seed AGUGCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302c-3p.2
|
|
|
hsa-miR-520f-3p
|
MIMAT0002830 |
Activity profile of AGUGCUU motif
Sorted Z-values of AGUGCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_235491462 | 0.37 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr11_-_102962929 | 0.34 |
ENST00000260247.5
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr18_+_9136758 | 0.27 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr18_-_18691739 | 0.23 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr3_+_30648066 | 0.22 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr12_+_19592602 | 0.21 |
ENST00000398864.3
ENST00000266508.9 |
AEBP2
|
AE binding protein 2 |
| chr2_+_174219548 | 0.21 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chrX_-_83442915 | 0.20 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr7_-_28220354 | 0.19 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr8_-_53626974 | 0.18 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr12_+_20522179 | 0.18 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr4_+_174089904 | 0.17 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr1_-_36235529 | 0.16 |
ENST00000318121.3
ENST00000373220.3 ENST00000520551.1 |
CLSPN
|
claspin |
| chr11_+_58346584 | 0.15 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr3_+_179065474 | 0.15 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr2_+_173940442 | 0.15 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr2_+_198380289 | 0.14 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr2_+_118846008 | 0.14 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr7_+_77325738 | 0.14 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr13_-_79233314 | 0.14 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr12_+_104458235 | 0.14 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr20_+_8112824 | 0.14 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr17_-_38083843 | 0.14 |
ENST00000304046.2
ENST00000579695.1 |
ORMDL3
|
ORM1-like 3 (S. cerevisiae) |
| chr12_+_56137064 | 0.13 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr6_-_111136513 | 0.13 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr4_-_53525406 | 0.13 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr20_+_33814457 | 0.12 |
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted) |
| chr12_-_46766577 | 0.12 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr9_-_123476719 | 0.12 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chrX_-_20284958 | 0.12 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr22_+_30279144 | 0.12 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr21_-_27542972 | 0.12 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr5_-_32444828 | 0.12 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr17_+_5185552 | 0.12 |
ENST00000262477.6
ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr1_+_42846443 | 0.11 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr11_-_77532050 | 0.11 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr6_+_157099036 | 0.11 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr3_+_88188254 | 0.11 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr9_+_108210279 | 0.11 |
ENST00000374716.4
ENST00000374710.3 ENST00000481272.1 ENST00000484973.1 ENST00000394926.3 ENST00000539376.1 |
FSD1L
|
fibronectin type III and SPRY domain containing 1-like |
| chr17_+_44928946 | 0.11 |
ENST00000290015.2
ENST00000393461.2 |
WNT9B
|
wingless-type MMTV integration site family, member 9B |
| chr6_-_108395907 | 0.11 |
ENST00000193322.3
|
OSTM1
|
osteopetrosis associated transmembrane protein 1 |
| chr13_+_97874574 | 0.11 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_-_99395787 | 0.11 |
ENST00000369244.2
ENST00000229971.1 |
FBXL4
|
F-box and leucine-rich repeat protein 4 |
| chr1_+_33207381 | 0.11 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr10_+_112631547 | 0.11 |
ENST00000280154.7
ENST00000393104.2 |
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr4_+_99916765 | 0.11 |
ENST00000296411.6
|
METAP1
|
methionyl aminopeptidase 1 |
| chr12_+_45609893 | 0.10 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chr16_-_57318566 | 0.10 |
ENST00000569059.1
ENST00000219207.5 |
PLLP
|
plasmolipin |
| chr8_-_103876965 | 0.10 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr12_-_65146636 | 0.10 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr20_+_44462430 | 0.10 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr1_-_173991434 | 0.10 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr11_-_94964354 | 0.10 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chrX_+_41192595 | 0.10 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr12_-_42538657 | 0.09 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr3_-_125094093 | 0.09 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr18_-_72921303 | 0.09 |
ENST00000322342.3
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr1_-_78148324 | 0.09 |
ENST00000370801.3
ENST00000433749.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr12_-_31744031 | 0.09 |
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr3_-_149093499 | 0.09 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr6_+_18155560 | 0.09 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr1_-_205180664 | 0.09 |
ENST00000367161.3
ENST00000367162.3 ENST00000367160.4 |
DSTYK
|
dual serine/threonine and tyrosine protein kinase |
| chr1_+_89149905 | 0.09 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr13_-_30424821 | 0.08 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr17_-_5138099 | 0.08 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr20_+_30865429 | 0.08 |
ENST00000375712.3
|
KIF3B
|
kinesin family member 3B |
| chr7_+_99613195 | 0.08 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr8_+_58907104 | 0.08 |
ENST00000361488.3
|
FAM110B
|
family with sequence similarity 110, member B |
| chr11_-_27494279 | 0.08 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_+_112162381 | 0.08 |
ENST00000433097.1
ENST00000369709.3 ENST00000436150.2 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr2_+_203499901 | 0.08 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr11_-_119599794 | 0.08 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr2_+_178257372 | 0.07 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr16_-_66730583 | 0.07 |
ENST00000330687.4
ENST00000394106.2 ENST00000563952.1 |
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr17_-_79919154 | 0.07 |
ENST00000409678.3
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chrX_-_20134990 | 0.07 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chr3_-_56835967 | 0.07 |
ENST00000495373.1
ENST00000296315.3 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr1_+_214776516 | 0.07 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr6_-_90121938 | 0.07 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr19_-_2050852 | 0.07 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr15_+_38544476 | 0.07 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr11_+_32914579 | 0.07 |
ENST00000399302.2
|
QSER1
|
glutamine and serine rich 1 |
| chr8_+_82644669 | 0.07 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr10_+_94050913 | 0.07 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr4_-_121993673 | 0.07 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
| chr12_-_76953284 | 0.07 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_-_70883708 | 0.07 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr3_-_155572164 | 0.07 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr3_-_176914238 | 0.06 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr2_+_103236004 | 0.06 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr1_-_65432171 | 0.06 |
ENST00000342505.4
|
JAK1
|
Janus kinase 1 |
| chr1_-_161039456 | 0.06 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_-_197169672 | 0.06 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr2_+_220110177 | 0.06 |
ENST00000409638.3
ENST00000396738.2 ENST00000409516.3 |
STK16
|
serine/threonine kinase 16 |
| chr5_+_133861790 | 0.06 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr6_+_14117872 | 0.06 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr1_+_203595903 | 0.06 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_-_85780086 | 0.06 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr9_-_125693757 | 0.06 |
ENST00000373656.3
|
ZBTB26
|
zinc finger and BTB domain containing 26 |
| chr10_-_61666267 | 0.06 |
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6 |
| chr1_+_61547894 | 0.06 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr5_+_139027877 | 0.06 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr11_-_67980744 | 0.06 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr6_-_32095968 | 0.06 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr1_-_57045228 | 0.05 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr11_-_110167352 | 0.05 |
ENST00000533991.1
ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX
|
radixin |
| chr16_-_47495170 | 0.05 |
ENST00000320640.6
ENST00000544001.2 |
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr4_-_102268628 | 0.05 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_+_122526383 | 0.05 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr13_+_39612442 | 0.05 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr6_+_158244223 | 0.05 |
ENST00000392185.3
|
SNX9
|
sorting nexin 9 |
| chr15_-_91537723 | 0.05 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr21_-_40685477 | 0.05 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr9_+_137218362 | 0.05 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr10_+_85899196 | 0.05 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr7_-_74867509 | 0.05 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr1_-_100598444 | 0.05 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chrX_-_70288234 | 0.05 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr13_-_22033392 | 0.05 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr7_+_139026057 | 0.05 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr7_-_2354099 | 0.05 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr14_-_75593708 | 0.05 |
ENST00000557673.1
ENST00000238616.5 |
NEK9
|
NIMA-related kinase 9 |
| chr1_+_87380299 | 0.05 |
ENST00000370551.4
ENST00000370550.5 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr15_+_76135622 | 0.04 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr9_-_130639997 | 0.04 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr9_-_86571628 | 0.04 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr11_+_61520075 | 0.04 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr5_-_159827073 | 0.04 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr16_+_69221028 | 0.04 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr3_-_69435224 | 0.04 |
ENST00000398540.3
|
FRMD4B
|
FERM domain containing 4B |
| chr12_+_50344516 | 0.04 |
ENST00000199280.3
ENST00000550862.1 |
AQP2
|
aquaporin 2 (collecting duct) |
| chr1_+_213123915 | 0.04 |
ENST00000366968.4
ENST00000490792.1 |
VASH2
|
vasohibin 2 |
| chr11_+_35684288 | 0.04 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr9_-_111929560 | 0.04 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr20_+_5986727 | 0.04 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr6_+_149887377 | 0.04 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chrX_+_108780062 | 0.04 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_-_86948245 | 0.04 |
ENST00000439940.2
ENST00000604011.1 |
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3 RNF103-CHMP3 readthrough |
| chr12_+_51442101 | 0.04 |
ENST00000550929.1
ENST00000262055.4 ENST00000550442.1 ENST00000549340.1 ENST00000548209.1 ENST00000548251.1 ENST00000550814.1 ENST00000547660.1 ENST00000380123.2 ENST00000548401.1 ENST00000418425.2 ENST00000547008.1 ENST00000552739.1 |
LETMD1
|
LETM1 domain containing 1 |
| chr12_+_88536067 | 0.04 |
ENST00000549011.1
ENST00000266712.6 ENST00000551088.1 |
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr10_+_1095416 | 0.04 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chrX_-_99986494 | 0.04 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr10_-_61469837 | 0.04 |
ENST00000395348.3
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr14_+_57046500 | 0.04 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr13_-_25746416 | 0.04 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr22_-_36236265 | 0.04 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_+_25071848 | 0.04 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr17_+_56270084 | 0.04 |
ENST00000225371.5
|
EPX
|
eosinophil peroxidase |
| chr17_-_71088797 | 0.04 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr4_-_114900831 | 0.04 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr1_+_93913713 | 0.04 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr10_+_98741041 | 0.04 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr4_-_99851766 | 0.03 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr6_-_3457256 | 0.03 |
ENST00000436008.2
|
SLC22A23
|
solute carrier family 22, member 23 |
| chr15_-_56535464 | 0.03 |
ENST00000559447.2
ENST00000422057.1 ENST00000317318.6 ENST00000423270.1 |
RFX7
|
regulatory factor X, 7 |
| chr11_-_105892937 | 0.03 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr7_+_139044621 | 0.03 |
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chr6_-_116381918 | 0.03 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chrX_+_77166172 | 0.03 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr11_-_96076334 | 0.03 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr9_+_19408999 | 0.03 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr1_-_70671216 | 0.03 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr1_-_205601064 | 0.03 |
ENST00000357992.4
ENST00000289703.4 |
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1) |
| chr7_+_104654623 | 0.03 |
ENST00000311117.3
ENST00000334877.4 ENST00000257745.4 ENST00000334914.7 ENST00000478990.1 ENST00000495267.1 ENST00000476671.1 |
KMT2E
|
lysine (K)-specific methyltransferase 2E |
| chr10_-_73976025 | 0.03 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr20_+_56884752 | 0.03 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr16_-_3493528 | 0.03 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr5_-_32313019 | 0.03 |
ENST00000280285.5
ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr2_-_105946491 | 0.03 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr17_-_1083078 | 0.03 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chrX_+_16737718 | 0.03 |
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1 |
| chr8_-_103668114 | 0.03 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr2_+_26568965 | 0.03 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr11_+_64126614 | 0.03 |
ENST00000528057.1
ENST00000334205.4 ENST00000294261.4 |
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr5_+_170814803 | 0.03 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr1_-_150552006 | 0.03 |
ENST00000307940.3
ENST00000369026.2 |
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related) |
| chr1_+_97187318 | 0.03 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr1_-_85156216 | 0.03 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr9_+_116638562 | 0.03 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr9_-_110251836 | 0.03 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr1_+_78245303 | 0.03 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr11_+_92085262 | 0.03 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr10_+_180987 | 0.03 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr22_-_41252962 | 0.03 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr2_+_170683942 | 0.03 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr17_-_2304365 | 0.03 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr1_+_26438289 | 0.03 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr12_-_95467356 | 0.03 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr3_-_53381539 | 0.03 |
ENST00000606822.1
ENST00000294241.6 ENST00000607628.1 |
DCP1A
|
decapping mRNA 1A |
| chr2_+_27593389 | 0.03 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr17_+_46125707 | 0.03 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr6_+_13615554 | 0.03 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr6_-_56707943 | 0.03 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr11_+_9406169 | 0.02 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr3_+_152017181 | 0.02 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr3_+_42695176 | 0.02 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr12_+_56915713 | 0.02 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr17_-_8868991 | 0.02 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr17_-_62658186 | 0.02 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
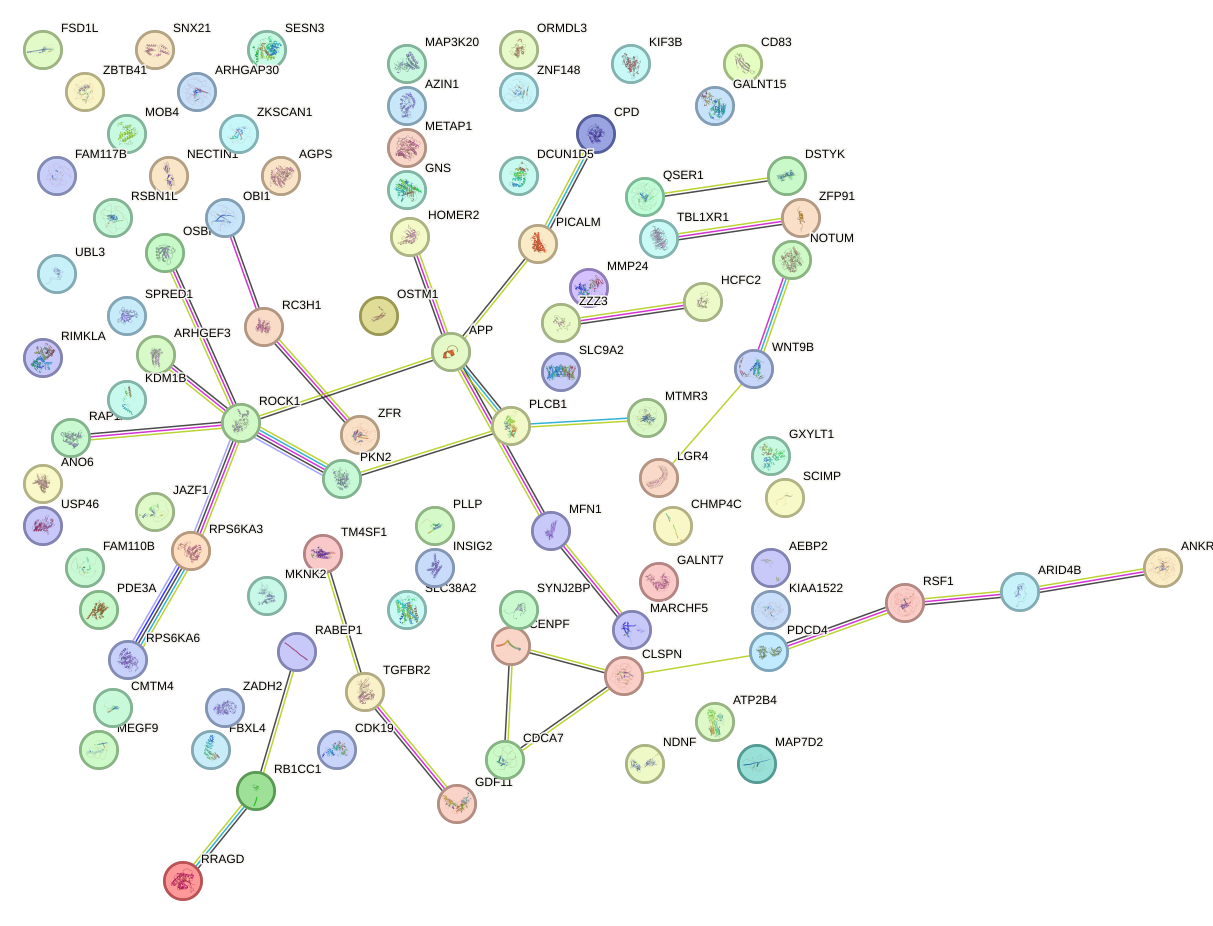
Network of associatons between targets according to the STRING database.
First level regulatory network of AGUGCUU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.1 | GO:0098736 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0072190 | ureter urothelium development(GO:0072190) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:2000301 | response to antineoplastic agent(GO:0097327) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0016939 | plus-end kinesin complex(GO:0005873) kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |