Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ALX3
Z-value: 0.69
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | 0.97 | 2.8e-02 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_20187174 | 0.33 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr4_+_95128748 | 0.32 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_66536248 | 0.31 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr12_-_25348007 | 0.28 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr14_+_57671888 | 0.27 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr2_+_13677795 | 0.27 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr3_+_138340067 | 0.26 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_+_107738343 | 0.23 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr4_+_71859156 | 0.23 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr11_-_13011081 | 0.21 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chr9_-_5830768 | 0.20 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr13_+_111748183 | 0.19 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr17_-_57229155 | 0.19 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_+_100862811 | 0.19 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr13_+_40229764 | 0.19 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chr16_+_31724618 | 0.19 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr9_-_75488984 | 0.19 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr2_-_136678123 | 0.18 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr16_+_72088376 | 0.18 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chrX_+_114827851 | 0.18 |
ENST00000539310.1
|
PLS3
|
plastin 3 |
| chr1_+_81771806 | 0.18 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr21_-_39705323 | 0.17 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr1_-_150669500 | 0.17 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_-_54411240 | 0.16 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_-_10022735 | 0.16 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr10_+_90660832 | 0.16 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr12_+_28410128 | 0.16 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr12_+_64798095 | 0.16 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr14_+_55493920 | 0.16 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr15_+_96904487 | 0.16 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr2_+_103089756 | 0.15 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chrX_+_43515467 | 0.15 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr15_+_64680003 | 0.15 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr6_+_28317685 | 0.15 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr5_+_68860949 | 0.14 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr11_-_107729504 | 0.14 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr5_-_94890648 | 0.14 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr1_+_186265399 | 0.14 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr17_-_39341594 | 0.14 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr12_-_74686314 | 0.14 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr6_-_82957433 | 0.13 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr4_-_25865159 | 0.13 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_+_90674665 | 0.13 |
ENST00000549313.1
|
RP11-753N8.1
|
RP11-753N8.1 |
| chr5_-_98262240 | 0.13 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr4_-_74486109 | 0.13 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_102200948 | 0.13 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_-_107729287 | 0.13 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr6_+_37897735 | 0.13 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr1_+_28261533 | 0.13 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chrX_+_108779004 | 0.13 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr9_-_115095123 | 0.13 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr11_+_101918153 | 0.12 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr10_+_35894338 | 0.12 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chr1_-_158173659 | 0.12 |
ENST00000415019.1
|
RP11-404O13.5
|
RP11-404O13.5 |
| chr18_-_53303123 | 0.12 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr10_+_24755416 | 0.12 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr16_+_31724552 | 0.12 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr20_-_56265680 | 0.12 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_28261492 | 0.12 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_62521614 | 0.11 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr6_+_26440700 | 0.11 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr3_+_148709128 | 0.11 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr8_-_93978333 | 0.11 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr14_+_55494323 | 0.11 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr1_-_207226313 | 0.11 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr17_-_64187973 | 0.11 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chr11_+_57480046 | 0.11 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr4_-_85654615 | 0.11 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr2_-_152118276 | 0.10 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr12_-_10978957 | 0.10 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr16_+_31885079 | 0.10 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr1_-_202897724 | 0.10 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr4_-_103749105 | 0.10 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_162336686 | 0.10 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr8_-_30670384 | 0.10 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr5_+_179135246 | 0.10 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr13_-_86373536 | 0.10 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr12_+_16500037 | 0.10 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr16_-_28621353 | 0.10 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr8_-_90769422 | 0.10 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr4_-_89442940 | 0.10 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr6_-_7313381 | 0.10 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr1_-_54411255 | 0.10 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr7_-_111424506 | 0.10 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr1_+_28261621 | 0.09 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_-_103748880 | 0.09 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_2883650 | 0.09 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr9_-_21482312 | 0.09 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr12_-_10282742 | 0.09 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_+_156433573 | 0.09 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr4_-_103749313 | 0.08 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_140482926 | 0.08 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr9_+_90112767 | 0.08 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr13_+_76413852 | 0.08 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr10_+_91152303 | 0.08 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr17_-_12920994 | 0.08 |
ENST00000609101.1
|
ELAC2
|
elaC ribonuclease Z 2 |
| chr17_+_18086392 | 0.08 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr16_+_53133070 | 0.08 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_5710919 | 0.08 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr9_-_99540328 | 0.08 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr1_-_150669604 | 0.08 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_+_16500599 | 0.08 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr8_-_124741451 | 0.08 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr1_+_157963063 | 0.08 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr7_-_44580861 | 0.08 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr3_-_57260377 | 0.08 |
ENST00000495160.2
|
HESX1
|
HESX homeobox 1 |
| chr6_-_127780510 | 0.08 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr20_+_57594309 | 0.07 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr2_+_11682790 | 0.07 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr1_+_151735431 | 0.07 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr15_-_75748115 | 0.07 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr7_+_138145076 | 0.07 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr1_+_66820058 | 0.07 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr11_+_100784231 | 0.07 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr2_+_187371440 | 0.07 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr5_+_66300446 | 0.07 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_16500571 | 0.07 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chrX_+_114827818 | 0.07 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr4_-_103749179 | 0.07 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_231860596 | 0.07 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr10_-_28571015 | 0.07 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr15_+_58724184 | 0.07 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr17_-_19364269 | 0.07 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr15_-_37393406 | 0.07 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr5_+_115177178 | 0.07 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr17_-_19015945 | 0.07 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr7_-_92777606 | 0.06 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_-_24306835 | 0.06 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr14_+_67831576 | 0.06 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr8_-_12612962 | 0.06 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr19_-_36001113 | 0.06 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr4_-_185275104 | 0.06 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr19_-_14064114 | 0.06 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr10_-_90611566 | 0.06 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr5_+_140201183 | 0.06 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr17_-_64225508 | 0.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr6_+_4087664 | 0.06 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr12_-_118797475 | 0.06 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_26087509 | 0.06 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chrX_+_84258832 | 0.05 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr2_-_152118352 | 0.05 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr7_-_87856280 | 0.05 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr7_+_37723450 | 0.05 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_-_87342564 | 0.05 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr9_+_90112117 | 0.05 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr9_+_90112590 | 0.05 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr8_-_93978309 | 0.05 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr16_+_15489629 | 0.05 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr3_-_160823040 | 0.05 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr8_-_623547 | 0.05 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr11_-_559377 | 0.05 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chrX_+_77166172 | 0.05 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr9_+_90112741 | 0.04 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr4_+_113568207 | 0.04 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_84630645 | 0.04 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chrX_+_56100757 | 0.04 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chr6_+_52442083 | 0.04 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr17_+_40610862 | 0.04 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr14_-_67878917 | 0.04 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr2_+_102953608 | 0.04 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr12_-_46121554 | 0.04 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr7_-_111424462 | 0.04 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chrX_-_71458802 | 0.04 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr1_+_220267429 | 0.04 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr4_-_120243545 | 0.04 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr22_+_42834029 | 0.04 |
ENST00000428765.1
|
CTA-126B4.7
|
CTA-126B4.7 |
| chr9_-_4666421 | 0.04 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr15_+_75970672 | 0.04 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr12_+_122688090 | 0.04 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr11_+_35201826 | 0.04 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr10_+_6821545 | 0.04 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr1_+_207226574 | 0.03 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr16_+_15489603 | 0.03 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr1_+_225600404 | 0.03 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr12_+_104337515 | 0.03 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr8_-_29605625 | 0.03 |
ENST00000506121.3
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr8_+_107738240 | 0.03 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr3_+_40518599 | 0.03 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr4_-_122686261 | 0.03 |
ENST00000337677.5
|
TMEM155
|
transmembrane protein 155 |
| chr10_+_70661014 | 0.03 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr2_-_3521518 | 0.03 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr12_-_10282836 | 0.03 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_-_77225135 | 0.03 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr11_+_29181503 | 0.03 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr11_+_118398178 | 0.03 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr8_-_93978357 | 0.03 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr3_-_126327398 | 0.03 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_-_101925055 | 0.03 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr6_-_32095968 | 0.03 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr19_+_17638041 | 0.03 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr7_+_99202003 | 0.03 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr19_+_42817527 | 0.03 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr6_+_26087646 | 0.03 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr6_+_34204642 | 0.02 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr17_-_38821373 | 0.02 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr10_+_695888 | 0.02 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr6_+_127898312 | 0.02 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chrX_-_102510126 | 0.02 |
ENST00000372685.3
|
TCEAL8
|
transcription elongation factor A (SII)-like 8 |
| chr12_+_59194154 | 0.02 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
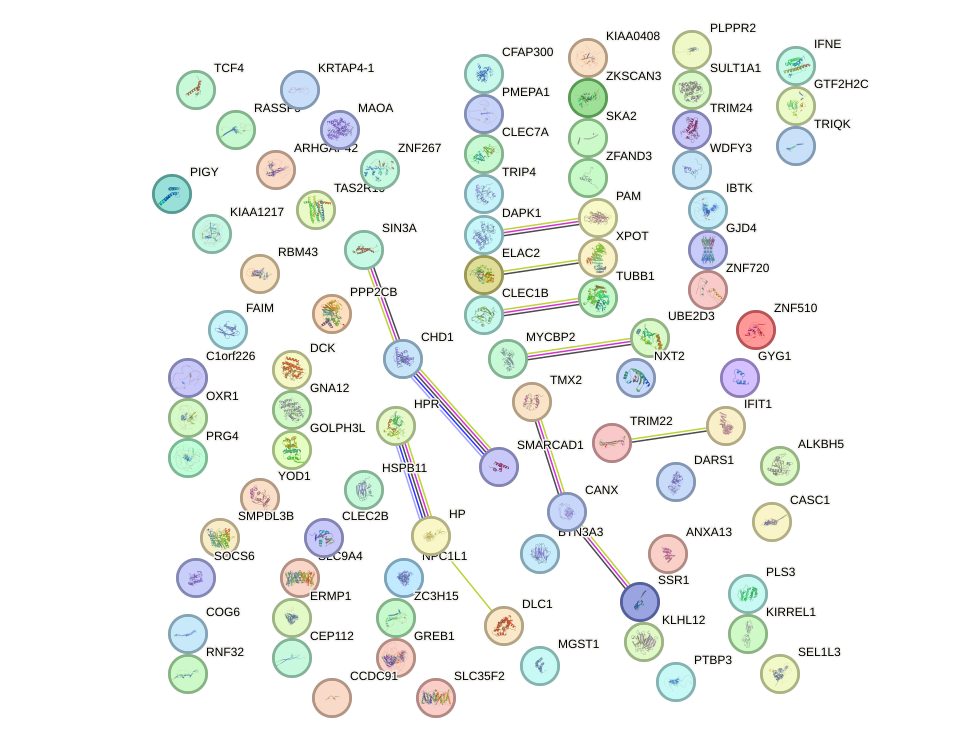
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.0 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |