Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
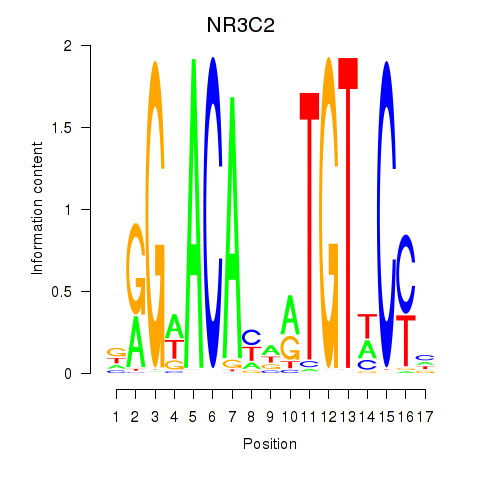
Results for AR_NR3C2
Z-value: 0.59
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AR
|
ENSG00000169083.11 | androgen receptor |
|
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AR | hg19_v2_chrX_+_66764375_66764465 | -0.90 | 9.6e-02 | Click! |
| NR3C2 | hg19_v2_chr4_-_149363662_149363688 | -0.63 | 3.7e-01 | Click! |
Activity profile of AR_NR3C2 motif
Sorted Z-values of AR_NR3C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_56642041 | 1.55 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr16_+_56642489 | 1.26 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr16_+_56716336 | 0.83 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr11_+_18287801 | 0.61 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr1_-_1149506 | 0.59 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr11_+_18287721 | 0.54 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr19_-_42916499 | 0.44 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr13_+_114462193 | 0.35 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr9_-_106087316 | 0.33 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr7_-_75241096 | 0.33 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr1_+_1846519 | 0.31 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr8_-_49833978 | 0.30 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr20_+_45947246 | 0.28 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr22_+_18632666 | 0.25 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr14_-_24616426 | 0.25 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr19_+_35630926 | 0.23 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35741466 | 0.23 |
ENST00000599658.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr8_-_49834299 | 0.21 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr6_+_127898312 | 0.21 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr15_+_31658349 | 0.19 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr8_-_72756667 | 0.19 |
ENST00000325509.4
|
MSC
|
musculin |
| chr19_+_35630628 | 0.18 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr14_+_24616588 | 0.16 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr1_-_145715565 | 0.15 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr8_-_21669826 | 0.15 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr19_+_57874835 | 0.15 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr4_-_185395191 | 0.14 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr11_-_114271139 | 0.14 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr11_+_48002076 | 0.13 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr8_-_145634666 | 0.13 |
ENST00000531042.1
|
CPSF1
|
cleavage and polyadenylation specific factor 1, 160kDa |
| chr4_-_69817481 | 0.12 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr6_+_123317116 | 0.12 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr10_+_26932068 | 0.12 |
ENST00000544033.1
ENST00000454991.1 |
LINC00202-2
RP13-16H11.7
|
long intergenic non-protein coding RNA 202-2 RP13-16H11.7 |
| chr4_-_135122903 | 0.12 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr12_+_122241928 | 0.11 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr2_-_75788424 | 0.11 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr4_+_75311019 | 0.11 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr19_+_35630344 | 0.10 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr14_-_106845789 | 0.10 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr11_-_5959840 | 0.10 |
ENST00000412903.1
|
TRIM5
|
tripartite motif containing 5 |
| chr1_+_93811438 | 0.10 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr11_+_65851443 | 0.09 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr9_+_131683174 | 0.09 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr12_-_106480587 | 0.09 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr16_-_4852915 | 0.09 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr2_+_238395879 | 0.09 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr19_+_1261106 | 0.09 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr12_+_66582919 | 0.09 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr1_-_204119009 | 0.08 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr2_+_238395803 | 0.08 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr11_+_103907308 | 0.08 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr4_+_75310851 | 0.08 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr3_-_119396193 | 0.08 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr12_+_1099675 | 0.08 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr5_+_140186647 | 0.08 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_-_10856694 | 0.07 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr14_-_106866934 | 0.07 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chrX_-_117119243 | 0.07 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr19_-_3786354 | 0.07 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr1_-_49242553 | 0.07 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr14_-_23623577 | 0.07 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr10_-_375422 | 0.07 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr22_+_19939026 | 0.07 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase |
| chr18_-_48346415 | 0.06 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr12_-_10251603 | 0.06 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr16_-_25269134 | 0.06 |
ENST00000328086.7
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2 |
| chr1_+_76262552 | 0.06 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr19_-_3786253 | 0.06 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_150147711 | 0.06 |
ENST00000307271.3
|
GIMAP8
|
GTPase, IMAP family member 8 |
| chrX_-_80377162 | 0.06 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr4_-_8442438 | 0.06 |
ENST00000356406.5
ENST00000413009.2 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr17_-_19290117 | 0.06 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr2_-_231825668 | 0.06 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr19_+_8117636 | 0.06 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr2_+_27435179 | 0.06 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr2_+_242577097 | 0.05 |
ENST00000419606.1
ENST00000474739.2 ENST00000396411.3 ENST00000425239.1 ENST00000400771.3 ENST00000430617.2 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr8_-_145634731 | 0.05 |
ENST00000349769.3
|
CPSF1
|
cleavage and polyadenylation specific factor 1, 160kDa |
| chr2_+_242577027 | 0.05 |
ENST00000402096.1
ENST00000404914.3 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr4_+_75480629 | 0.05 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr19_-_14117074 | 0.05 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr11_-_75380165 | 0.05 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chrX_+_2609207 | 0.04 |
ENST00000381192.3
|
CD99
|
CD99 molecule |
| chr14_-_106692191 | 0.04 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_-_24469602 | 0.04 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr17_+_38498594 | 0.04 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chr19_-_8408139 | 0.04 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr4_-_69215699 | 0.04 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr1_-_209979465 | 0.04 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chr8_+_91013577 | 0.04 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr16_-_787728 | 0.04 |
ENST00000567403.1
ENST00000562421.1 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr6_-_131299929 | 0.03 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_106668095 | 0.03 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr18_+_47087055 | 0.03 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr2_+_27435734 | 0.03 |
ENST00000419744.1
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr12_+_51318513 | 0.03 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr3_-_58563094 | 0.03 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr19_+_8117881 | 0.03 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr3_+_118864991 | 0.03 |
ENST00000295622.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chrX_+_2609317 | 0.03 |
ENST00000381187.3
ENST00000381184.1 |
CD99
|
CD99 molecule |
| chr2_-_242576864 | 0.02 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr17_+_79031415 | 0.02 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr3_-_98235962 | 0.02 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr1_-_27693349 | 0.02 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr11_-_506739 | 0.02 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr16_-_66584059 | 0.02 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_+_118865028 | 0.02 |
ENST00000460150.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chr22_+_38054721 | 0.02 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr15_-_28344439 | 0.02 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
| chr14_-_106552755 | 0.02 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chrX_+_2609356 | 0.02 |
ENST00000381180.3
ENST00000449611.1 |
CD99
|
CD99 molecule |
| chr7_-_4877677 | 0.02 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr19_+_58570605 | 0.01 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr8_-_110615669 | 0.01 |
ENST00000533394.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr17_+_38333263 | 0.01 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr19_-_45657028 | 0.01 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr19_+_54704610 | 0.01 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr17_-_16472483 | 0.01 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr12_+_56615761 | 0.01 |
ENST00000447747.1
ENST00000399713.2 |
NABP2
|
nucleic acid binding protein 2 |
| chr16_+_67207872 | 0.01 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr16_+_67207838 | 0.00 |
ENST00000566871.1
ENST00000268605.7 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr11_-_236326 | 0.00 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr8_+_9046503 | 0.00 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr11_+_236540 | 0.00 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr2_-_109605663 | 0.00 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of AR_NR3C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.8 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.5 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 1.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0050666 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 3.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |