Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CAGUCCA
Z-value: 0.27
miRNA associated with seed CAGUCCA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-455-3p.1
|
MIMAT0004784 |
Activity profile of CAGUCCA motif
Sorted Z-values of CAGUCCA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_203274639 | 0.15 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr4_+_174089904 | 0.14 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr1_-_146644122 | 0.14 |
ENST00000254101.3
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr8_-_91658303 | 0.11 |
ENST00000458549.2
|
TMEM64
|
transmembrane protein 64 |
| chr2_-_55237484 | 0.10 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr6_-_29527702 | 0.10 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr14_-_100070363 | 0.09 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr15_+_49170083 | 0.09 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr5_+_65018017 | 0.09 |
ENST00000380985.5
ENST00000502464.1 |
NLN
|
neurolysin (metallopeptidase M3 family) |
| chr5_+_60628074 | 0.09 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr3_-_37034702 | 0.08 |
ENST00000322716.5
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1 |
| chr1_-_54304212 | 0.08 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr1_+_201979645 | 0.08 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr3_+_124223586 | 0.08 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr9_+_103204553 | 0.08 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr8_-_103876965 | 0.08 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr5_+_67511524 | 0.08 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_111136513 | 0.08 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr19_+_47421933 | 0.07 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr15_+_96873921 | 0.07 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_46671323 | 0.07 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr5_+_92919043 | 0.07 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr9_-_104145795 | 0.06 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr17_-_48943706 | 0.06 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr19_+_8274204 | 0.06 |
ENST00000561053.1
ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4
|
ceramide synthase 4 |
| chr6_-_10415470 | 0.06 |
ENST00000379604.2
ENST00000379613.3 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr9_+_103235365 | 0.06 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_-_150979333 | 0.05 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr8_-_91997427 | 0.05 |
ENST00000517562.2
|
RP11-122A3.2
|
chromosome 8 open reading frame 88 |
| chr13_+_88324870 | 0.05 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr12_-_90049828 | 0.05 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_+_41952591 | 0.05 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr4_-_99851766 | 0.05 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr2_-_61765315 | 0.05 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr11_+_14665263 | 0.05 |
ENST00000282096.4
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited |
| chr16_-_18937726 | 0.05 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_+_53848505 | 0.05 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr1_-_40105617 | 0.05 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr4_+_124320665 | 0.05 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr9_+_100745615 | 0.05 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr19_+_45973120 | 0.05 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr2_-_172290482 | 0.04 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr14_-_53258314 | 0.04 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chrX_+_16804544 | 0.04 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr6_+_147830063 | 0.04 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr10_+_98592009 | 0.04 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr4_-_100871506 | 0.04 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr1_+_153700518 | 0.04 |
ENST00000318967.2
ENST00000456435.1 ENST00000435409.2 |
INTS3
|
integrator complex subunit 3 |
| chr2_+_48010221 | 0.04 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr3_-_182698381 | 0.04 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr2_+_157291953 | 0.04 |
ENST00000310454.6
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr1_+_27153173 | 0.04 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr5_+_137673945 | 0.04 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr17_+_56160768 | 0.04 |
ENST00000579991.2
|
DYNLL2
|
dynein, light chain, LC8-type 2 |
| chr10_-_60027642 | 0.04 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr11_-_129062093 | 0.04 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_+_22224811 | 0.04 |
ENST00000381237.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_75988321 | 0.03 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr16_+_22217577 | 0.03 |
ENST00000263026.5
|
EEF2K
|
eukaryotic elongation factor-2 kinase |
| chr14_-_64010046 | 0.03 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr15_+_45315302 | 0.03 |
ENST00000267814.9
|
SORD
|
sorbitol dehydrogenase |
| chr12_+_54447637 | 0.03 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr17_+_57697216 | 0.03 |
ENST00000393043.1
ENST00000269122.3 |
CLTC
|
clathrin, heavy chain (Hc) |
| chr12_-_27091183 | 0.03 |
ENST00000544548.1
ENST00000261191.7 ENST00000537336.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr15_+_41851211 | 0.03 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr4_-_71705590 | 0.03 |
ENST00000254799.6
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr11_-_45307817 | 0.03 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr14_+_57857262 | 0.02 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr20_+_306221 | 0.02 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr1_-_40367530 | 0.02 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr8_+_26149007 | 0.02 |
ENST00000380737.3
ENST00000524169.1 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr1_-_33338076 | 0.02 |
ENST00000496770.1
|
FNDC5
|
fibronectin type III domain containing 5 |
| chr10_-_71930222 | 0.02 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr15_+_76135622 | 0.02 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr11_-_118305921 | 0.02 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr1_-_17766198 | 0.02 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr11_+_125439298 | 0.02 |
ENST00000278903.6
ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24
|
etoposide induced 2.4 |
| chr12_+_68042495 | 0.02 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr3_+_196466710 | 0.02 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chrX_+_133507327 | 0.02 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr13_-_50367057 | 0.02 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr8_+_22409193 | 0.02 |
ENST00000240123.7
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chrX_+_48554986 | 0.02 |
ENST00000376687.3
ENST00000453214.2 |
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr21_+_47878757 | 0.02 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr15_+_49715293 | 0.02 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr17_-_41977964 | 0.02 |
ENST00000377184.3
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr14_+_23775971 | 0.02 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr3_+_25469724 | 0.02 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr9_+_115513003 | 0.02 |
ENST00000374232.3
|
SNX30
|
sorting nexin family member 30 |
| chr13_-_45915221 | 0.02 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr16_-_46865047 | 0.02 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_+_9773409 | 0.02 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr2_-_197036289 | 0.02 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr11_-_82782861 | 0.02 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr17_-_16395455 | 0.01 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr17_-_48227877 | 0.01 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr5_-_159546396 | 0.01 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr19_+_19496624 | 0.01 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr11_-_134281812 | 0.01 |
ENST00000392580.1
ENST00000312527.4 |
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr16_+_12995468 | 0.01 |
ENST00000424107.3
ENST00000558583.1 ENST00000558318.1 |
SHISA9
|
shisa family member 9 |
| chr10_+_88516396 | 0.01 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chrX_+_69664706 | 0.01 |
ENST00000194900.4
ENST00000374360.3 |
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr3_-_11762202 | 0.01 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chrX_+_153639856 | 0.01 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chr17_-_7120525 | 0.01 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr5_-_76935513 | 0.01 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr7_+_69064300 | 0.01 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr2_-_73298802 | 0.01 |
ENST00000411783.1
ENST00000410065.1 ENST00000442582.1 ENST00000272433.2 |
SFXN5
|
sideroflexin 5 |
| chr7_+_73242069 | 0.01 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr15_+_44719394 | 0.01 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr3_-_132004251 | 0.01 |
ENST00000512055.1
|
CPNE4
|
copine IV |
| chr12_-_71148413 | 0.01 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_85360499 | 0.01 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr10_-_33246722 | 0.01 |
ENST00000437302.1
ENST00000396033.2 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr20_-_656823 | 0.01 |
ENST00000246104.6
|
SCRT2
|
scratch family zinc finger 2 |
| chr3_+_183903811 | 0.01 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr15_+_90931450 | 0.01 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chr2_+_61404624 | 0.01 |
ENST00000394457.3
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr5_+_56111361 | 0.01 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr3_-_128902729 | 0.01 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr2_+_27440229 | 0.01 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr14_+_58666824 | 0.01 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr1_+_75594119 | 0.01 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr3_+_38495333 | 0.01 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr9_+_4679555 | 0.01 |
ENST00000381858.1
ENST00000381854.3 |
CDC37L1
|
cell division cycle 37-like 1 |
| chr8_-_30670384 | 0.01 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr6_-_17706618 | 0.01 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr2_+_232651124 | 0.00 |
ENST00000350033.3
ENST00000412591.1 ENST00000410017.1 ENST00000373608.3 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr2_+_105471969 | 0.00 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chrX_-_134049262 | 0.00 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chrX_-_25034065 | 0.00 |
ENST00000379044.4
|
ARX
|
aristaless related homeobox |
| chr7_-_152133059 | 0.00 |
ENST00000262189.6
ENST00000355193.2 |
KMT2C
|
lysine (K)-specific methyltransferase 2C |
| chr6_-_90529418 | 0.00 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr2_-_131850951 | 0.00 |
ENST00000409185.1
ENST00000389915.3 |
FAM168B
|
family with sequence similarity 168, member B |
| chr15_-_100273544 | 0.00 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr1_-_213189108 | 0.00 |
ENST00000535388.1
|
ANGEL2
|
angel homolog 2 (Drosophila) |
| chr12_+_98987369 | 0.00 |
ENST00000401722.3
ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| chr18_-_29522989 | 0.00 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
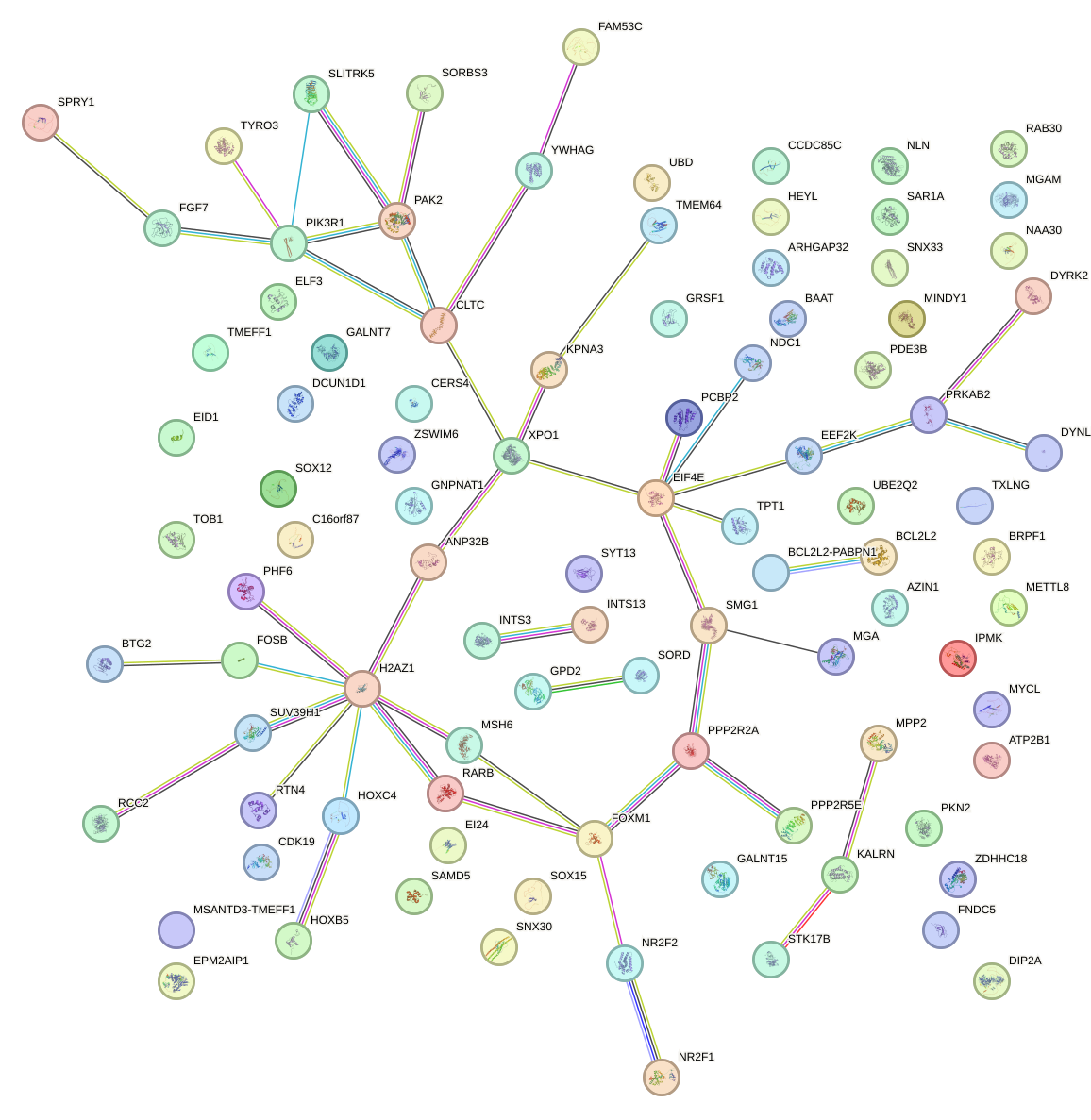
Network of associatons between targets according to the STRING database.
First level regulatory network of CAGUCCA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.0 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |