Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GLIS2
Z-value: 1.47
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4382225_4382225 | -0.40 | 6.0e-01 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_667759 | 1.05 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr18_-_59274139 | 0.81 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr19_+_38880695 | 0.78 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_-_123634449 | 0.76 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr2_+_177001685 | 0.69 |
ENST00000432796.2
|
HOXD3
|
homeobox D3 |
| chr7_-_130080818 | 0.64 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr1_-_156675564 | 0.63 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_41132088 | 0.61 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr15_-_90233907 | 0.61 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr10_-_9801179 | 0.55 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chr12_+_57854274 | 0.54 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr10_-_14880566 | 0.53 |
ENST00000378442.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr10_+_14880364 | 0.52 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr22_-_21213676 | 0.51 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr3_+_118892411 | 0.50 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr19_+_22817119 | 0.50 |
ENST00000456783.2
|
ZNF492
|
zinc finger protein 492 |
| chr20_+_55108302 | 0.49 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr19_-_2282167 | 0.48 |
ENST00000342063.3
|
C19orf35
|
chromosome 19 open reading frame 35 |
| chr19_+_4304685 | 0.47 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr19_+_45973360 | 0.47 |
ENST00000589593.1
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr18_+_12948000 | 0.46 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr19_-_39303576 | 0.45 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr5_+_149865377 | 0.45 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_-_153304697 | 0.45 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr19_-_17007026 | 0.44 |
ENST00000598792.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr6_-_168397757 | 0.40 |
ENST00000456585.1
ENST00000414364.1 |
KIF25-AS1
|
KIF25 antisense RNA 1 |
| chr12_+_122018697 | 0.39 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr2_-_230786378 | 0.39 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr1_-_153348067 | 0.38 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr13_-_20767037 | 0.37 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr14_+_101293687 | 0.37 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_-_61197406 | 0.37 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr19_+_39926791 | 0.37 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr1_+_186649754 | 0.36 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr6_-_153304148 | 0.36 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr2_-_27718052 | 0.36 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr8_+_98881268 | 0.36 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr8_-_95565673 | 0.36 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr17_-_27467418 | 0.35 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_627143 | 0.35 |
ENST00000176195.3
|
SCT
|
secretin |
| chr8_+_118533049 | 0.35 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr9_+_135037334 | 0.34 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr3_+_153839149 | 0.34 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr19_-_51071302 | 0.34 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr1_-_54411255 | 0.34 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr3_-_16554403 | 0.34 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr19_-_50979981 | 0.34 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr2_+_170655789 | 0.33 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr8_+_104426942 | 0.33 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr1_-_156675368 | 0.33 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr9_-_115095229 | 0.32 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr9_+_139847347 | 0.32 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr12_+_57998595 | 0.32 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr3_+_28390637 | 0.32 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr1_-_40157345 | 0.32 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr9_+_26956474 | 0.32 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr20_-_30433396 | 0.32 |
ENST00000375978.3
|
FOXS1
|
forkhead box S1 |
| chr10_-_124768300 | 0.32 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr3_-_52719912 | 0.31 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr2_+_46769798 | 0.31 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr15_-_77712429 | 0.31 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr10_+_114709999 | 0.31 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_1356628 | 0.31 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr6_-_97731019 | 0.31 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr16_-_84587633 | 0.31 |
ENST00000565079.1
|
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr1_+_87595433 | 0.31 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr16_+_333152 | 0.30 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr11_+_31531291 | 0.30 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr17_-_4463856 | 0.30 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr12_+_57998400 | 0.30 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr20_-_23402028 | 0.29 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr1_-_22215192 | 0.29 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr12_-_49110613 | 0.29 |
ENST00000261900.3
|
CCNT1
|
cyclin T1 |
| chr3_-_197282821 | 0.29 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_+_5389605 | 0.29 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr22_-_50699972 | 0.28 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr14_-_50999307 | 0.28 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr11_+_14665263 | 0.28 |
ENST00000282096.4
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited |
| chr19_+_2249308 | 0.28 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr2_+_242716231 | 0.27 |
ENST00000192314.6
|
GAL3ST2
|
galactose-3-O-sulfotransferase 2 |
| chr14_-_92302784 | 0.27 |
ENST00000340892.5
ENST00000360594.5 |
TC2N
|
tandem C2 domains, nuclear |
| chr21_-_39288743 | 0.27 |
ENST00000609713.1
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr6_-_39282221 | 0.26 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr21_-_44751903 | 0.26 |
ENST00000450205.1
|
LINC00322
|
long intergenic non-protein coding RNA 322 |
| chr11_+_393428 | 0.26 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr2_-_45162783 | 0.26 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr19_-_55866104 | 0.26 |
ENST00000326529.4
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr7_+_150758642 | 0.26 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr2_-_46769694 | 0.26 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr3_-_52719888 | 0.26 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr6_+_96969672 | 0.26 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr17_-_45056606 | 0.25 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr1_-_242162375 | 0.25 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr1_+_213224572 | 0.25 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr5_+_68788594 | 0.25 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr2_-_55277692 | 0.25 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr15_+_42694573 | 0.25 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr3_-_11888246 | 0.25 |
ENST00000455809.1
ENST00000444133.2 ENST00000273037.5 |
TAMM41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chr22_-_46931191 | 0.24 |
ENST00000454637.1
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr5_-_169816638 | 0.24 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr20_-_52687059 | 0.24 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr11_+_68451943 | 0.24 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr5_+_133707252 | 0.24 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr16_-_89752965 | 0.24 |
ENST00000567544.1
|
RP11-368I7.4
|
Uncharacterized protein |
| chr20_+_52824367 | 0.23 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr19_-_45661995 | 0.23 |
ENST00000438936.2
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr3_-_193096600 | 0.23 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr6_-_41673552 | 0.23 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr13_-_114018400 | 0.23 |
ENST00000375430.4
ENST00000375431.4 |
GRTP1
|
growth hormone regulated TBC protein 1 |
| chr5_-_141060389 | 0.23 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr14_-_54418598 | 0.23 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr7_+_2695726 | 0.23 |
ENST00000429448.1
|
TTYH3
|
tweety family member 3 |
| chr11_-_76381029 | 0.22 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr14_+_103566481 | 0.22 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr11_-_64510409 | 0.22 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr3_-_28390581 | 0.22 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr2_+_24272576 | 0.22 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr2_+_71357744 | 0.22 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr17_-_58469329 | 0.22 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr7_+_77167376 | 0.22 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr1_-_16939976 | 0.21 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr19_+_24269981 | 0.21 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chrX_+_53449887 | 0.21 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr1_-_115632035 | 0.21 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr16_-_32688053 | 0.21 |
ENST00000398682.4
|
TP53TG3
|
TP53 target 3 |
| chr1_-_27701307 | 0.20 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr11_+_86749035 | 0.20 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr6_-_133119668 | 0.20 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr20_-_52687030 | 0.20 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr13_-_108867846 | 0.20 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr1_+_65613217 | 0.19 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr8_-_11325047 | 0.19 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr6_-_28367510 | 0.19 |
ENST00000361028.1
|
ZSCAN12
|
zinc finger and SCAN domain containing 12 |
| chr12_+_132568814 | 0.19 |
ENST00000454179.2
ENST00000389560.2 ENST00000443539.2 ENST00000392352.1 |
EP400NL
|
EP400 N-terminal like |
| chr6_+_144164455 | 0.19 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr4_+_37828255 | 0.19 |
ENST00000381967.4
ENST00000544359.1 ENST00000537241.1 |
PGM2
|
phosphoglucomutase 2 |
| chr3_+_151531810 | 0.19 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr13_-_22033392 | 0.19 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr9_+_26956371 | 0.19 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr14_-_105531759 | 0.19 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr6_+_2988933 | 0.19 |
ENST00000597787.1
|
LINC01011
|
long intergenic non-protein coding RNA 1011 |
| chr1_+_35544968 | 0.18 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr19_-_41196534 | 0.18 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr8_-_29120604 | 0.18 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr6_+_17393839 | 0.18 |
ENST00000489374.1
ENST00000378990.2 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr17_-_36760865 | 0.18 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr17_+_61571746 | 0.18 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr10_+_14880287 | 0.18 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr17_+_38278530 | 0.18 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr6_+_138725343 | 0.18 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr17_-_34079897 | 0.18 |
ENST00000254466.6
ENST00000587565.1 |
GAS2L2
|
growth arrest-specific 2 like 2 |
| chr14_-_23426231 | 0.18 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr13_-_108867101 | 0.18 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr14_+_100150622 | 0.17 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr11_+_86748957 | 0.17 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr2_-_171571077 | 0.17 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr2_-_179343268 | 0.17 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr17_-_56065484 | 0.17 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_+_181845074 | 0.17 |
ENST00000602959.1
ENST00000602479.1 ENST00000392415.2 ENST00000602291.1 |
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr2_-_38604398 | 0.17 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr19_+_45973120 | 0.17 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr7_-_138363824 | 0.17 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr19_-_39108643 | 0.17 |
ENST00000396857.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr19_+_45409011 | 0.17 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr3_-_49933249 | 0.16 |
ENST00000434765.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr3_+_44626446 | 0.16 |
ENST00000441021.1
ENST00000322734.2 |
ZNF660
|
zinc finger protein 660 |
| chr20_-_52210368 | 0.16 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr12_-_95467356 | 0.16 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chrX_+_53449805 | 0.16 |
ENST00000414955.2
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr3_-_63849571 | 0.16 |
ENST00000295899.5
|
THOC7
|
THO complex 7 homolog (Drosophila) |
| chr4_-_106394866 | 0.16 |
ENST00000502596.1
|
PPA2
|
pyrophosphatase (inorganic) 2 |
| chr12_-_95467267 | 0.16 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr1_+_91966656 | 0.16 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr19_+_45254529 | 0.15 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr2_+_131113609 | 0.15 |
ENST00000347849.3
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr19_-_3772209 | 0.15 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr1_-_226187013 | 0.15 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr6_-_114292449 | 0.15 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr7_-_130080681 | 0.15 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr17_-_4269768 | 0.15 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr11_-_117695449 | 0.15 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr11_-_72353494 | 0.15 |
ENST00000544570.1
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_-_55277654 | 0.15 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr1_+_150954493 | 0.15 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr19_+_58570605 | 0.15 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr4_-_107237340 | 0.15 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase |
| chr4_+_107237660 | 0.14 |
ENST00000394701.4
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr11_+_86748863 | 0.14 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr9_+_125795788 | 0.14 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr1_-_201084796 | 0.14 |
ENST00000458416.1
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr17_+_18012020 | 0.14 |
ENST00000205890.5
|
MYO15A
|
myosin XVA |
| chr11_-_1619524 | 0.14 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr2_+_131113580 | 0.14 |
ENST00000175756.5
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr7_+_2685164 | 0.14 |
ENST00000400376.2
|
TTYH3
|
tweety family member 3 |
| chr19_-_55791431 | 0.14 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_+_38498594 | 0.14 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chr6_-_168720425 | 0.13 |
ENST00000366796.3
|
DACT2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr19_+_56116771 | 0.13 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr19_-_44124019 | 0.13 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr19_-_19006890 | 0.13 |
ENST00000247005.6
|
GDF1
|
growth differentiation factor 1 |
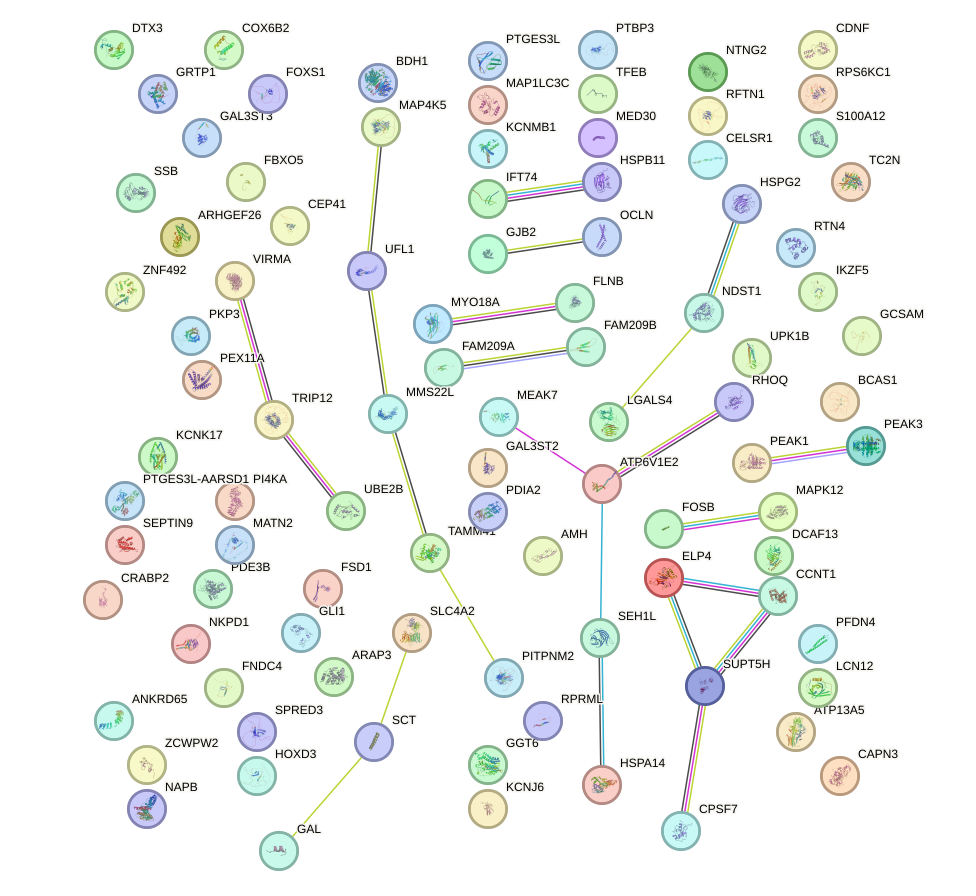
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.4 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 0.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.2 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.0 | 0.9 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.2 | GO:0061155 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.2 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.6 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0090140 | response to food(GO:0032094) regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.3 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |