Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
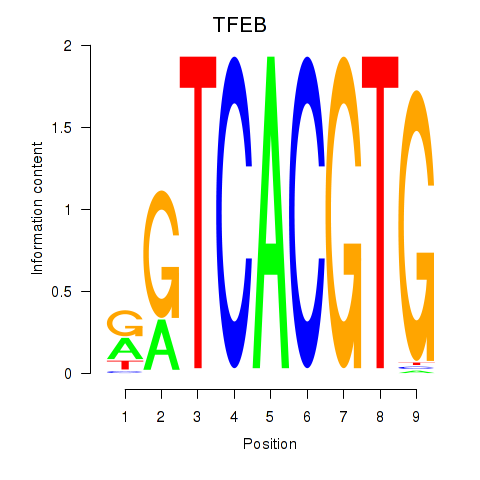
Results for MAX_TFEB
Z-value: 0.58
Transcription factors associated with MAX_TFEB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAX
|
ENSG00000125952.14 | MYC associated factor X |
|
TFEB
|
ENSG00000112561.13 | transcription factor EB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFEB | hg19_v2_chr6_-_41673552_41673678 | 0.36 | 6.4e-01 | Click! |
| MAX | hg19_v2_chr14_-_65569057_65569119 | -0.32 | 6.8e-01 | Click! |
Activity profile of MAX_TFEB motif
Sorted Z-values of MAX_TFEB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58145889 | 2.52 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr12_+_123849462 | 0.49 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr15_+_44084503 | 0.44 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr9_-_131709858 | 0.36 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr15_+_44084040 | 0.34 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr19_+_49458107 | 0.31 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr14_-_81687197 | 0.30 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr6_+_138188351 | 0.30 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_-_58146128 | 0.29 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr19_-_10764509 | 0.29 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr12_-_58146048 | 0.29 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr20_-_44519839 | 0.28 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr19_+_10764937 | 0.28 |
ENST00000449870.1
ENST00000318511.3 ENST00000420083.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr17_-_7137582 | 0.27 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr6_+_139349903 | 0.27 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr14_-_81687575 | 0.23 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr19_+_1275917 | 0.23 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr17_-_7137857 | 0.23 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr3_+_133292851 | 0.23 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr10_-_76995675 | 0.20 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr12_+_56110247 | 0.20 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_+_8386371 | 0.20 |
ENST00000600659.2
|
RPS28
|
ribosomal protein S28 |
| chr17_-_48450534 | 0.20 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr19_-_5680891 | 0.20 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr9_+_131709966 | 0.20 |
ENST00000372577.2
|
NUP188
|
nucleoporin 188kDa |
| chr9_-_100684845 | 0.19 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr17_+_6915798 | 0.19 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr16_-_1525016 | 0.19 |
ENST00000262318.8
ENST00000448525.1 |
CLCN7
|
chloride channel, voltage-sensitive 7 |
| chr2_+_240323439 | 0.18 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr1_-_11865982 | 0.18 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr12_+_54332535 | 0.18 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chr1_+_150254936 | 0.18 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr19_+_4402659 | 0.17 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr13_+_113951532 | 0.17 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr17_+_6915730 | 0.17 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr15_-_83315874 | 0.17 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_+_41256764 | 0.17 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr20_+_44519948 | 0.17 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr6_-_33385870 | 0.17 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_-_61560053 | 0.17 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr16_+_2570431 | 0.17 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr17_+_6915902 | 0.17 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr12_+_56109926 | 0.16 |
ENST00000547076.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr9_-_100684769 | 0.16 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr19_+_10812108 | 0.16 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr17_-_6915646 | 0.16 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr11_+_67159416 | 0.15 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr14_-_20929624 | 0.15 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr21_-_46237883 | 0.15 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr16_-_28503357 | 0.15 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr3_+_51428704 | 0.15 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr12_-_49463753 | 0.15 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr16_-_28503080 | 0.15 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr5_-_1799932 | 0.15 |
ENST00000382647.7
ENST00000505059.2 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr12_+_56109810 | 0.15 |
ENST00000550412.1
ENST00000257899.2 ENST00000548925.1 ENST00000549147.1 |
RP11-644F5.10
BLOC1S1
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_+_10765003 | 0.14 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr10_-_76995769 | 0.14 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr20_+_44520009 | 0.14 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chr19_+_7587491 | 0.14 |
ENST00000264079.6
|
MCOLN1
|
mucolipin 1 |
| chr19_-_4670345 | 0.14 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr11_-_61560254 | 0.14 |
ENST00000543510.1
|
TMEM258
|
transmembrane protein 258 |
| chrX_+_118108571 | 0.14 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr17_-_73851285 | 0.14 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr6_-_33385823 | 0.14 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385655 | 0.14 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr16_+_2014941 | 0.13 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr19_-_11545920 | 0.13 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr17_-_7197881 | 0.13 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr6_-_33385854 | 0.13 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_46970178 | 0.13 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_62223320 | 0.13 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr14_-_20923195 | 0.13 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr16_+_84178874 | 0.13 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr12_+_56110315 | 0.13 |
ENST00000548556.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_-_10047219 | 0.13 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr17_-_48450265 | 0.12 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr19_+_54609230 | 0.12 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr16_-_2097787 | 0.12 |
ENST00000566380.1
ENST00000219066.1 |
NTHL1
|
nth endonuclease III-like 1 (E. coli) |
| chr4_+_76439095 | 0.12 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr1_+_44440575 | 0.12 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr4_+_164265035 | 0.12 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr12_-_90103077 | 0.12 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_22778337 | 0.12 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr14_-_64761078 | 0.12 |
ENST00000341099.4
ENST00000556275.1 ENST00000542956.1 ENST00000353772.3 ENST00000357782.2 ENST00000267525.6 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr2_-_133427767 | 0.12 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr7_+_916183 | 0.12 |
ENST00000265857.3
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr16_+_88636789 | 0.12 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr3_-_49395705 | 0.12 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr16_+_1401924 | 0.12 |
ENST00000204679.4
ENST00000529110.1 |
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr11_+_118938485 | 0.11 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr3_+_50654821 | 0.11 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr22_+_35776828 | 0.11 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chrX_-_100662881 | 0.11 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr5_-_1799965 | 0.11 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr1_-_212873267 | 0.11 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr11_-_61646054 | 0.11 |
ENST00000527379.1
|
FADS3
|
fatty acid desaturase 3 |
| chr20_+_2821366 | 0.11 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr22_-_24622080 | 0.11 |
ENST00000425408.1
|
GGT5
|
gamma-glutamyltransferase 5 |
| chr2_+_198365095 | 0.11 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr11_-_18343725 | 0.11 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr8_-_123793048 | 0.11 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr14_+_77564701 | 0.10 |
ENST00000557115.1
|
KIAA1737
|
CLOCK-interacting pacemaker |
| chr16_-_28503327 | 0.10 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_+_2098003 | 0.10 |
ENST00000439673.2
ENST00000350773.4 |
TSC2
|
tuberous sclerosis 2 |
| chr22_-_30661807 | 0.10 |
ENST00000403389.1
|
OSM
|
oncostatin M |
| chr6_-_44225231 | 0.10 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr1_+_11866207 | 0.10 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr17_-_37886752 | 0.10 |
ENST00000577810.1
|
MIEN1
|
migration and invasion enhancer 1 |
| chr19_-_48018203 | 0.10 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr11_-_61684962 | 0.10 |
ENST00000394836.2
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr7_-_27205136 | 0.10 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr19_-_5720248 | 0.10 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr1_+_26758849 | 0.10 |
ENST00000533087.1
ENST00000531312.1 ENST00000525165.1 ENST00000525326.1 ENST00000525546.1 ENST00000436153.2 ENST00000530781.1 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr19_+_5681153 | 0.10 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr17_+_35306175 | 0.10 |
ENST00000225402.5
|
AATF
|
apoptosis antagonizing transcription factor |
| chr11_+_62538775 | 0.10 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr14_-_93799360 | 0.10 |
ENST00000334746.5
ENST00000554565.1 ENST00000298896.3 |
BTBD7
|
BTB (POZ) domain containing 7 |
| chr18_-_3247084 | 0.10 |
ENST00000609924.1
|
RP13-270P17.3
|
RP13-270P17.3 |
| chr16_+_28985542 | 0.10 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr12_+_117176113 | 0.10 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr15_+_41913690 | 0.10 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr11_-_36310958 | 0.10 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr1_+_11866270 | 0.10 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr17_+_79935418 | 0.10 |
ENST00000306729.7
ENST00000306739.4 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr5_+_176730769 | 0.10 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr2_+_232575168 | 0.10 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr1_-_33116128 | 0.10 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr15_+_32322685 | 0.10 |
ENST00000454250.3
ENST00000306901.3 |
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr3_-_142720267 | 0.09 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr17_+_42977122 | 0.09 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr8_-_145688231 | 0.09 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chrX_+_102840408 | 0.09 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr16_+_28986134 | 0.09 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr16_-_88923285 | 0.09 |
ENST00000542788.1
ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr16_-_18937072 | 0.09 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr17_+_48450575 | 0.09 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr1_-_100643765 | 0.09 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr3_+_133292759 | 0.09 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr19_+_14544099 | 0.09 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr1_+_32712815 | 0.09 |
ENST00000373582.3
|
FAM167B
|
family with sequence similarity 167, member B |
| chrX_+_118108601 | 0.09 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr6_-_31864977 | 0.09 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr20_+_2821340 | 0.09 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr17_+_79935464 | 0.09 |
ENST00000581647.1
ENST00000580534.1 ENST00000579684.1 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr6_+_7107999 | 0.09 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr11_-_8892900 | 0.09 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr3_+_183967409 | 0.09 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr19_-_8386238 | 0.09 |
ENST00000301457.2
|
NDUFA7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr1_+_221051699 | 0.09 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr11_+_130184888 | 0.09 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr9_-_90589586 | 0.09 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr9_+_130860810 | 0.09 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr10_+_51827648 | 0.09 |
ENST00000351071.6
ENST00000314664.7 ENST00000282633.5 |
FAM21A
|
family with sequence similarity 21, member A |
| chr2_+_198365122 | 0.09 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr7_+_100464760 | 0.09 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr19_+_7587555 | 0.09 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr12_-_104359475 | 0.09 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr16_+_57023406 | 0.09 |
ENST00000262510.6
ENST00000308149.7 ENST00000436936.1 |
NLRC5
|
NLR family, CARD domain containing 5 |
| chr16_+_4897632 | 0.09 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr11_+_64781575 | 0.09 |
ENST00000246747.4
ENST00000529384.1 |
ARL2
|
ADP-ribosylation factor-like 2 |
| chr19_-_40324767 | 0.09 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chrX_+_129040094 | 0.09 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr1_-_111743285 | 0.09 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr17_+_37356528 | 0.09 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr8_-_103876340 | 0.09 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr4_-_47465666 | 0.09 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr10_+_46222648 | 0.08 |
ENST00000336378.4
ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C
|
family with sequence similarity 21, member C |
| chr2_-_200715834 | 0.08 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr6_-_43197189 | 0.08 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr19_+_1067271 | 0.08 |
ENST00000536472.1
ENST00000590214.1 |
HMHA1
|
histocompatibility (minor) HA-1 |
| chr17_+_7476136 | 0.08 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr1_-_52521831 | 0.08 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_+_133706865 | 0.08 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr3_-_170626418 | 0.08 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr19_-_10426663 | 0.08 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chr16_+_2570340 | 0.08 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr11_+_64781657 | 0.08 |
ENST00000533729.1
|
ARL2
|
ADP-ribosylation factor-like 2 |
| chr7_+_75024903 | 0.08 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr17_-_36904437 | 0.08 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr1_-_28559502 | 0.08 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr15_+_32322709 | 0.08 |
ENST00000455693.2
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr19_+_11546093 | 0.08 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr3_-_49395892 | 0.08 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr16_+_810728 | 0.08 |
ENST00000563941.1
ENST00000545450.2 ENST00000566549.1 |
MSLN
|
mesothelin |
| chr11_-_18343669 | 0.08 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chrX_-_100872911 | 0.08 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr16_+_5083950 | 0.08 |
ENST00000588623.1
|
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr11_+_57508825 | 0.08 |
ENST00000534355.1
|
C11orf31
|
chromosome 11 open reading frame 31 |
| chr7_-_100808843 | 0.08 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr4_-_83295103 | 0.08 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr7_-_100808394 | 0.08 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr11_-_58611957 | 0.08 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr4_-_100871506 | 0.08 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr20_-_61051026 | 0.08 |
ENST00000252997.2
|
GATA5
|
GATA binding protein 5 |
| chr17_+_37356586 | 0.08 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr11_+_777562 | 0.08 |
ENST00000530083.1
|
AP006621.5
|
Protein LOC100506518 |
| chr17_+_37356555 | 0.08 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr10_-_88126224 | 0.08 |
ENST00000327946.7
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr16_+_28986085 | 0.08 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr17_-_7218631 | 0.08 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAX_TFEB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.5 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.4 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.3 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.5 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.1 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.0 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |