Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
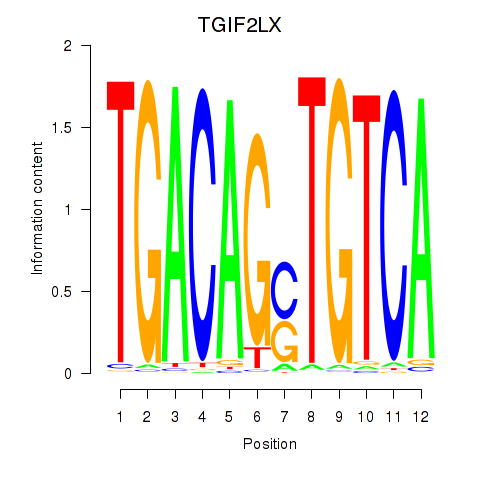
Results for MEIS3_TGIF2LX
Z-value: 0.87
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS3 | hg19_v2_chr19_-_47922750_47922795 | -0.94 | 6.3e-02 | Click! |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_42160486 | 1.56 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr1_+_79115503 | 0.90 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr11_-_615570 | 0.67 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr3_-_52719912 | 0.61 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr6_-_33663474 | 0.59 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr3_+_183899770 | 0.55 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr2_+_97202480 | 0.51 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr11_+_117947724 | 0.47 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr2_+_10560147 | 0.45 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr11_+_18287801 | 0.39 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr2_-_55920952 | 0.39 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr11_+_18287721 | 0.39 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr14_-_67859422 | 0.37 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr16_+_69458428 | 0.36 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr17_-_6917755 | 0.33 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr19_+_1000418 | 0.32 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr1_+_8378140 | 0.31 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr19_-_58609570 | 0.30 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr2_-_158182105 | 0.30 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr7_-_7782204 | 0.29 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr16_+_69458537 | 0.29 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr9_+_100174344 | 0.28 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr2_+_97203082 | 0.28 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr11_-_76381029 | 0.28 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr20_+_35504522 | 0.27 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr9_+_96026230 | 0.27 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chrX_-_153775426 | 0.26 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr1_+_46379254 | 0.26 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr1_+_159750720 | 0.26 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chrX_+_17755696 | 0.26 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr4_-_18023350 | 0.25 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr21_+_30503282 | 0.25 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr12_-_54978086 | 0.25 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr11_-_65325664 | 0.24 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_-_32264250 | 0.24 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr19_+_6361754 | 0.23 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr1_+_159750776 | 0.23 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr11_-_26743546 | 0.23 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_+_95611536 | 0.22 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr18_+_68002675 | 0.22 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr3_+_52719936 | 0.22 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr12_+_120875910 | 0.22 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr18_-_52989525 | 0.21 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr16_-_2004683 | 0.20 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr19_+_5623186 | 0.20 |
ENST00000538656.1
|
SAFB
|
scaffold attachment factor B |
| chr12_+_121647962 | 0.19 |
ENST00000542067.1
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr7_-_141957847 | 0.19 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr12_+_121647868 | 0.18 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr19_-_50400212 | 0.18 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr6_+_134273321 | 0.18 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chr2_-_24149918 | 0.18 |
ENST00000439915.1
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr20_-_3644046 | 0.18 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr2_-_175202151 | 0.17 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr15_+_65337708 | 0.16 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr10_-_7661623 | 0.16 |
ENST00000298441.6
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr1_-_47655686 | 0.16 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_+_228395755 | 0.16 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr5_+_155753745 | 0.16 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr16_+_30772913 | 0.16 |
ENST00000563909.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr16_+_81528948 | 0.15 |
ENST00000539778.2
|
CMIP
|
c-Maf inducing protein |
| chr19_-_4670345 | 0.15 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr2_+_181988560 | 0.14 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr9_-_130541017 | 0.14 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr9_-_113800705 | 0.14 |
ENST00000441240.1
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr17_-_9725388 | 0.14 |
ENST00000399363.4
|
RP11-477N12.3
|
Putative germ cell-specific gene 1-like protein 2 |
| chr3_+_122513642 | 0.14 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr7_+_29234375 | 0.13 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr14_-_68157084 | 0.13 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr2_-_179914760 | 0.13 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr8_+_99439214 | 0.13 |
ENST00000287042.4
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr16_-_69418553 | 0.13 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr19_+_18530146 | 0.13 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_-_842509 | 0.13 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr22_+_35776354 | 0.13 |
ENST00000412893.1
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr12_-_133787772 | 0.13 |
ENST00000545350.1
|
AC226150.4
|
Uncharacterized protein |
| chr1_+_155107820 | 0.13 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr9_+_137967268 | 0.13 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr7_-_128415844 | 0.13 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr7_-_48068643 | 0.12 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr6_-_30128657 | 0.12 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr1_+_210406121 | 0.12 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr18_+_3449695 | 0.12 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_145549203 | 0.12 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr17_+_34538310 | 0.11 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr18_+_55018044 | 0.11 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_-_34122596 | 0.11 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr6_-_32557610 | 0.11 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr6_+_31887761 | 0.11 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr22_-_31885514 | 0.11 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chrX_-_148713365 | 0.10 |
ENST00000511776.1
ENST00000507237.1 |
TMEM185A
|
transmembrane protein 185A |
| chr22_+_44319648 | 0.10 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr12_+_8850277 | 0.10 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr16_-_18468926 | 0.10 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr5_+_122110691 | 0.10 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr8_+_126104076 | 0.10 |
ENST00000517532.1
ENST00000287437.3 ENST00000518013.1 ENST00000522563.1 |
NSMCE2
|
non-SMC element 2, MMS21 homolog (S. cerevisiae) |
| chr1_-_32264356 | 0.10 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chrX_-_11445856 | 0.10 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr11_-_72145669 | 0.10 |
ENST00000543042.1
ENST00000294053.3 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr11_+_102188272 | 0.10 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr17_-_79533608 | 0.10 |
ENST00000572760.1
ENST00000573876.1 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr13_-_114312501 | 0.10 |
ENST00000335288.4
|
ATP4B
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chrX_+_41548220 | 0.09 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr2_+_27799389 | 0.09 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr2_+_233271546 | 0.09 |
ENST00000295453.3
|
ALPPL2
|
alkaline phosphatase, placental-like 2 |
| chr2_+_149402553 | 0.09 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr20_-_45142154 | 0.09 |
ENST00000347606.4
ENST00000457685.2 |
ZNF334
|
zinc finger protein 334 |
| chr1_-_54872059 | 0.09 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr17_+_40985407 | 0.09 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr5_+_115298165 | 0.09 |
ENST00000357872.4
|
AQPEP
|
Aminopeptidase Q |
| chr15_+_68924327 | 0.09 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr2_-_61697862 | 0.09 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr2_+_242089833 | 0.09 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr3_-_12883026 | 0.09 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr18_+_54318893 | 0.09 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr16_+_56899114 | 0.09 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr1_+_168250194 | 0.08 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr15_+_83478370 | 0.08 |
ENST00000286760.4
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr12_+_95611516 | 0.08 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_5680231 | 0.08 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr3_-_47023455 | 0.08 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr5_+_15500280 | 0.08 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr18_+_3449821 | 0.08 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr10_+_28822636 | 0.08 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr19_-_45826125 | 0.08 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr16_+_20817839 | 0.08 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr18_+_3449330 | 0.08 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr14_-_80677613 | 0.08 |
ENST00000556811.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr3_+_130745769 | 0.08 |
ENST00000412440.2
|
NEK11
|
NIMA-related kinase 11 |
| chr6_-_112194484 | 0.08 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr22_-_50051151 | 0.08 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr5_-_42887494 | 0.08 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_-_76381781 | 0.08 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr3_-_52719546 | 0.08 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr17_+_34431212 | 0.08 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr7_-_48068671 | 0.07 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr15_+_68115895 | 0.07 |
ENST00000554240.1
|
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr10_-_79398127 | 0.07 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_20817953 | 0.07 |
ENST00000568647.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr10_-_105212141 | 0.07 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr16_+_67360856 | 0.07 |
ENST00000568804.2
|
LRRC36
|
leucine rich repeat containing 36 |
| chr17_+_8316442 | 0.07 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr3_+_111260856 | 0.07 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr11_-_111749878 | 0.07 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr7_+_116654958 | 0.07 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_16539094 | 0.07 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr3_-_196987309 | 0.07 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_-_25801478 | 0.07 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr3_+_9691117 | 0.07 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr19_-_5680202 | 0.07 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chrX_-_13752675 | 0.07 |
ENST00000380579.1
ENST00000458511.2 ENST00000519885.1 ENST00000358231.5 ENST00000518847.1 ENST00000453655.2 ENST00000359680.5 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr10_-_105212059 | 0.07 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr11_+_7595136 | 0.07 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_-_74618907 | 0.07 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr5_+_166711804 | 0.07 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr2_-_239149300 | 0.07 |
ENST00000436051.1
|
HES6
|
hes family bHLH transcription factor 6 |
| chr9_-_140444814 | 0.06 |
ENST00000277531.4
|
PNPLA7
|
patatin-like phospholipase domain containing 7 |
| chr3_+_111260954 | 0.06 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr8_+_67039131 | 0.06 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr2_+_24714729 | 0.06 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr15_+_77287715 | 0.06 |
ENST00000559161.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr17_+_37782955 | 0.06 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr8_-_126103948 | 0.06 |
ENST00000523297.1
|
KIAA0196
|
KIAA0196 |
| chr22_+_46731596 | 0.06 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr5_+_173472607 | 0.06 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr21_-_46954529 | 0.06 |
ENST00000485649.2
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr6_+_53948328 | 0.06 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr15_+_85427903 | 0.05 |
ENST00000286749.3
ENST00000394573.1 ENST00000537703.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr18_+_54318616 | 0.05 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr12_+_95611569 | 0.05 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr3_-_64253655 | 0.05 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr1_+_144151520 | 0.05 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr4_-_134070250 | 0.05 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr6_+_151662815 | 0.05 |
ENST00000359755.5
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_+_111749650 | 0.05 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr19_-_5624057 | 0.05 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr13_-_41837620 | 0.05 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr19_+_17982747 | 0.05 |
ENST00000222248.3
|
SLC5A5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr11_+_102188224 | 0.05 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr11_-_31832581 | 0.05 |
ENST00000379111.2
|
PAX6
|
paired box 6 |
| chr2_+_110656005 | 0.05 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr16_+_20817926 | 0.05 |
ENST00000565340.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr11_+_128634589 | 0.05 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr19_+_6361795 | 0.05 |
ENST00000596149.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr16_+_20817746 | 0.05 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chrX_-_13835147 | 0.04 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr17_+_58755184 | 0.04 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr10_+_50507232 | 0.04 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr8_-_7243080 | 0.04 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr11_+_7273181 | 0.04 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chr11_-_111749767 | 0.04 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr11_-_107590383 | 0.04 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr17_-_42345487 | 0.04 |
ENST00000262418.6
|
SLC4A1
|
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr5_-_131329918 | 0.04 |
ENST00000357096.1
ENST00000431707.1 ENST00000434099.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr8_-_30890710 | 0.04 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr4_+_159727272 | 0.04 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chrX_+_23925918 | 0.04 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr1_-_57285038 | 0.04 |
ENST00000343433.6
|
C1orf168
|
chromosome 1 open reading frame 168 |
| chrX_-_23926004 | 0.04 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr11_+_63304273 | 0.04 |
ENST00000439013.2
ENST00000255688.3 |
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3 |
| chr19_-_10227503 | 0.04 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.4 | GO:2000625 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.2 | GO:1902460 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.4 | GO:0032308 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:1903823 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.8 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.6 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0045924 | regulation of female receptivity(GO:0045924) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |