Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SOX5
Z-value: 0.37
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SRY-box transcription factor 5 |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_78003204 | 0.26 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr6_-_134861089 | 0.19 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chrX_+_135252050 | 0.19 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135251783 | 0.18 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr7_+_129015671 | 0.16 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_+_129015484 | 0.15 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr5_-_146461027 | 0.13 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_+_135251835 | 0.13 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr12_+_78224667 | 0.12 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr17_+_40119801 | 0.12 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_-_138780159 | 0.12 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr2_-_145188137 | 0.11 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_89746264 | 0.11 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_+_41236325 | 0.11 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr7_-_111424506 | 0.11 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr12_-_71003568 | 0.11 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr9_-_98268883 | 0.10 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr17_+_67498295 | 0.10 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr21_+_35553045 | 0.10 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_+_71588372 | 0.10 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_-_200323414 | 0.10 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr7_-_111846435 | 0.10 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr1_-_12677714 | 0.10 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr7_+_77167343 | 0.10 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chrX_+_24483338 | 0.10 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr16_-_73093597 | 0.09 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr12_-_96793142 | 0.09 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr3_+_72201910 | 0.09 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr7_+_77167376 | 0.09 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_-_115375107 | 0.09 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr6_+_21666633 | 0.09 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr15_-_41166414 | 0.09 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr1_-_23340400 | 0.09 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr10_-_32667660 | 0.08 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr12_-_109797249 | 0.08 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr3_-_192635943 | 0.08 |
ENST00000392452.2
|
MB21D2
|
Mab-21 domain containing 2 |
| chr11_+_46402482 | 0.08 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr18_+_3451584 | 0.08 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_99873145 | 0.08 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chrX_+_123094369 | 0.08 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr3_+_19988736 | 0.08 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr8_-_128231299 | 0.08 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr17_-_46703826 | 0.08 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr4_+_160188889 | 0.08 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_133055815 | 0.08 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr11_+_126225529 | 0.08 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr12_+_60058458 | 0.08 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr18_+_3451646 | 0.07 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_+_28448995 | 0.07 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr18_+_21594585 | 0.07 |
ENST00000317571.3
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr15_+_66994663 | 0.07 |
ENST00000457357.2
|
SMAD6
|
SMAD family member 6 |
| chr10_-_14614311 | 0.07 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr12_+_78359999 | 0.07 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr1_+_156589198 | 0.07 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr7_+_106810165 | 0.07 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr20_-_45985414 | 0.07 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr7_-_83824449 | 0.07 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr20_-_45985172 | 0.07 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr9_-_74383302 | 0.07 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr14_-_73493825 | 0.06 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr20_-_45985464 | 0.06 |
ENST00000458360.2
ENST00000262975.4 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr5_+_138677515 | 0.06 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr5_+_115358088 | 0.06 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr18_-_52989525 | 0.06 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr16_+_20911174 | 0.06 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr6_+_89790459 | 0.06 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr15_+_66994561 | 0.06 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr5_-_38595498 | 0.06 |
ENST00000263409.4
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr6_+_89790490 | 0.06 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_-_158182322 | 0.06 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr7_-_7575477 | 0.06 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr2_-_158182105 | 0.06 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr3_+_171561127 | 0.06 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_228337553 | 0.06 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr12_+_9066472 | 0.06 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr10_+_121578211 | 0.06 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr10_-_104913367 | 0.06 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr11_-_82708519 | 0.05 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr3_+_89156799 | 0.05 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chrX_+_41192595 | 0.05 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr1_-_21503337 | 0.05 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_+_35996859 | 0.05 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr6_+_56954867 | 0.05 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr10_-_14614122 | 0.05 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_-_243418650 | 0.05 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr2_+_33661382 | 0.05 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_232573208 | 0.05 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr12_-_102874102 | 0.05 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr2_-_208030647 | 0.05 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_30569506 | 0.05 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chrY_+_15016013 | 0.05 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr10_+_97803151 | 0.05 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr15_-_34610962 | 0.05 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_-_158526756 | 0.05 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr3_-_65583561 | 0.05 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr12_-_89746173 | 0.05 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr5_+_138678131 | 0.05 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr11_-_46848393 | 0.05 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr10_-_14614095 | 0.05 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_+_164529004 | 0.04 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr6_+_126240442 | 0.04 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr2_+_27071045 | 0.04 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr11_-_72852320 | 0.04 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr2_+_232573222 | 0.04 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr2_+_58655461 | 0.04 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr9_-_92051391 | 0.04 |
ENST00000420681.2
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr6_-_13290684 | 0.04 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr7_+_29519662 | 0.04 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr18_+_20494078 | 0.04 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr11_-_115127611 | 0.04 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr6_-_49712123 | 0.04 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr7_-_111424462 | 0.04 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr6_-_133055896 | 0.04 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr9_+_137218362 | 0.04 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr14_-_92414294 | 0.04 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr11_-_64647144 | 0.04 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr14_-_69619823 | 0.04 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr2_-_26205340 | 0.04 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr14_-_69619291 | 0.04 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr1_-_39395165 | 0.04 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_-_73360796 | 0.04 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr11_+_110001723 | 0.04 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr12_-_51477333 | 0.04 |
ENST00000228515.1
ENST00000548206.1 ENST00000546935.1 ENST00000548981.1 |
CSRNP2
|
cysteine-serine-rich nuclear protein 2 |
| chr2_-_86422523 | 0.04 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr7_+_134576317 | 0.04 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr10_-_118897806 | 0.04 |
ENST00000277905.2
|
VAX1
|
ventral anterior homeobox 1 |
| chr14_-_73493784 | 0.04 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr17_-_56082455 | 0.04 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr19_-_7698599 | 0.04 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr12_-_8815404 | 0.04 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chrX_-_50557302 | 0.04 |
ENST00000289292.7
|
SHROOM4
|
shroom family member 4 |
| chr7_-_47622156 | 0.04 |
ENST00000457718.1
|
TNS3
|
tensin 3 |
| chr12_+_27623565 | 0.04 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr3_+_112930373 | 0.03 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr13_+_28813645 | 0.03 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr20_-_45984401 | 0.03 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr16_-_31085514 | 0.03 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr2_-_55277436 | 0.03 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr5_-_147162263 | 0.03 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr7_-_83824169 | 0.03 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr20_+_5987890 | 0.03 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr12_+_27398584 | 0.03 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr11_+_57435219 | 0.03 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr11_+_46402583 | 0.03 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_+_220495800 | 0.03 |
ENST00000413743.1
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr9_-_115249484 | 0.03 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr1_+_163038565 | 0.03 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr15_+_91416092 | 0.03 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_-_102874330 | 0.03 |
ENST00000307046.8
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr10_+_24528108 | 0.03 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chrX_-_53711064 | 0.03 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr19_+_40873617 | 0.03 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr12_-_111358372 | 0.03 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr2_+_203499901 | 0.03 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr9_-_37465396 | 0.03 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr2_-_55277512 | 0.03 |
ENST00000402434.2
|
RTN4
|
reticulon 4 |
| chr8_-_70745575 | 0.03 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr10_-_104953009 | 0.03 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chrX_-_50557014 | 0.03 |
ENST00000376020.2
|
SHROOM4
|
shroom family member 4 |
| chr12_-_12674032 | 0.03 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr11_-_89224638 | 0.03 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr12_-_110937351 | 0.03 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr2_-_160472952 | 0.03 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_-_43637915 | 0.03 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr10_+_35416090 | 0.03 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr9_+_132099158 | 0.03 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr2_-_55277654 | 0.03 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr8_-_116673894 | 0.03 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_+_110656268 | 0.03 |
ENST00000553749.1
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr6_+_100054606 | 0.03 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr6_+_155538093 | 0.03 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr3_-_129407535 | 0.03 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr6_-_32557610 | 0.03 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr10_-_62149433 | 0.03 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr6_-_49712072 | 0.03 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr9_-_92051354 | 0.03 |
ENST00000418828.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr11_-_79151695 | 0.03 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr14_-_23904861 | 0.03 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr20_-_45981138 | 0.03 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_+_25469802 | 0.03 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr3_+_19988566 | 0.03 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr13_-_46716969 | 0.02 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr19_-_19626351 | 0.02 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr3_+_134514093 | 0.02 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr1_+_11751748 | 0.02 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr3_-_11685345 | 0.02 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr12_-_498415 | 0.02 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr14_-_69619689 | 0.02 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr12_+_498545 | 0.02 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr12_-_6233828 | 0.02 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr7_+_54610124 | 0.02 |
ENST00000402026.2
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr11_+_36397528 | 0.02 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chrX_+_107288239 | 0.02 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_+_26435915 | 0.02 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr11_-_35440796 | 0.02 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_14613968 | 0.02 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr4_+_170581213 | 0.02 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr15_+_84115868 | 0.02 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr17_+_67498538 | 0.02 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_101439242 | 0.02 |
ENST00000296420.4
|
EMCN
|
endomucin |
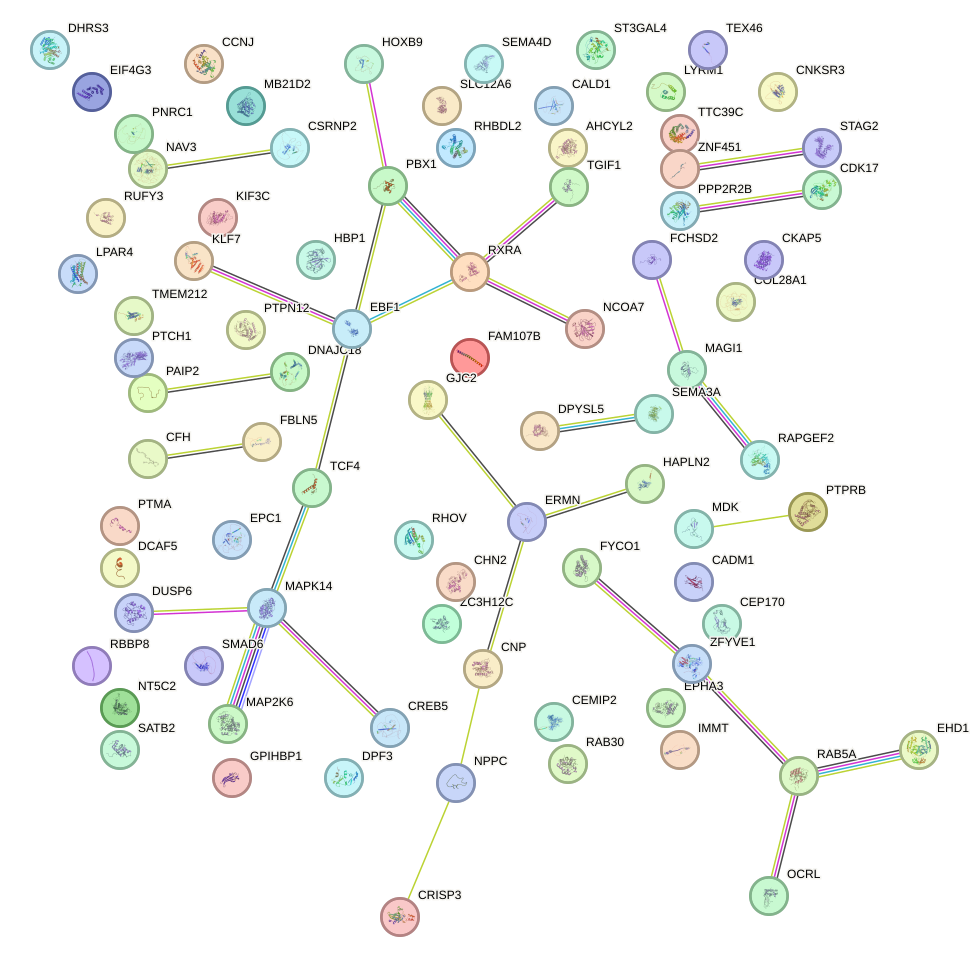
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.0 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.0 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0003183 | mitral valve morphogenesis(GO:0003183) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |