Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for THRB
Z-value: 0.81
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRB | hg19_v2_chr3_-_24536253_24536328 | 0.92 | 7.6e-02 | Click! |
Activity profile of THRB motif
Sorted Z-values of THRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32414059 | 2.09 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr10_+_62538248 | 0.99 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr20_+_3024266 | 0.97 |
ENST00000245983.2
ENST00000359100.2 ENST00000359987.1 |
GNRH2
|
gonadotropin-releasing hormone 2 |
| chr10_+_62538089 | 0.77 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr15_-_35838348 | 0.68 |
ENST00000561411.1
ENST00000256538.4 ENST00000440392.2 |
DPH6
|
diphthamine biosynthesis 6 |
| chr13_+_27844464 | 0.66 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr10_+_60145155 | 0.63 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr2_+_200472779 | 0.59 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr9_-_139268068 | 0.58 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr17_-_73840614 | 0.55 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr8_+_22102626 | 0.47 |
ENST00000519237.1
ENST00000397802.4 |
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr19_-_52643157 | 0.46 |
ENST00000597013.1
ENST00000600228.1 ENST00000596290.1 |
ZNF616
|
zinc finger protein 616 |
| chr8_+_22102611 | 0.44 |
ENST00000306433.4
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr11_-_45307817 | 0.38 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr8_-_131028782 | 0.36 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr17_+_1646130 | 0.34 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chrX_+_101854096 | 0.29 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr2_-_9563469 | 0.29 |
ENST00000484735.1
ENST00000456913.2 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr6_+_77484663 | 0.27 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr12_+_54378923 | 0.27 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr17_-_73839792 | 0.26 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr1_-_207224307 | 0.26 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr16_+_72088376 | 0.23 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr20_+_39969519 | 0.19 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr11_+_73498973 | 0.15 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr6_-_99842041 | 0.15 |
ENST00000254759.3
ENST00000369242.1 |
COQ3
|
coenzyme Q3 methyltransferase |
| chr1_-_154164534 | 0.14 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr1_-_113498616 | 0.13 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr17_-_73840774 | 0.13 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr2_-_9563216 | 0.13 |
ENST00000467606.1
ENST00000494563.1 ENST00000460001.1 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr11_-_47399942 | 0.12 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_+_28174649 | 0.11 |
ENST00000301908.3
|
PNOC
|
prepronociceptin |
| chr10_+_123970670 | 0.09 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_124037890 | 0.09 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr11_+_5617976 | 0.09 |
ENST00000445329.1
|
TRIM6
|
tripartite motif containing 6 |
| chr1_+_32712815 | 0.09 |
ENST00000373582.3
|
FAM167B
|
family with sequence similarity 167, member B |
| chr17_-_73840415 | 0.09 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr11_-_64014379 | 0.08 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_+_102937869 | 0.07 |
ENST00000249269.4
ENST00000428154.1 ENST00000420236.2 |
PMPCB
|
peptidase (mitochondrial processing) beta |
| chr11_-_47400078 | 0.07 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_-_113498943 | 0.07 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_-_47400062 | 0.07 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_-_9563319 | 0.05 |
ENST00000497105.1
ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr3_+_50606577 | 0.05 |
ENST00000434410.1
ENST00000232854.4 |
HEMK1
|
HemK methyltransferase family member 1 |
| chr11_+_5617858 | 0.03 |
ENST00000380097.3
|
TRIM6
|
tripartite motif containing 6 |
| chr5_+_131746575 | 0.03 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr9_+_100069933 | 0.02 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr11_+_46366918 | 0.02 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr11_+_46366799 | 0.01 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr8_+_19674651 | 0.01 |
ENST00000397977.3
|
INTS10
|
integrator complex subunit 10 |
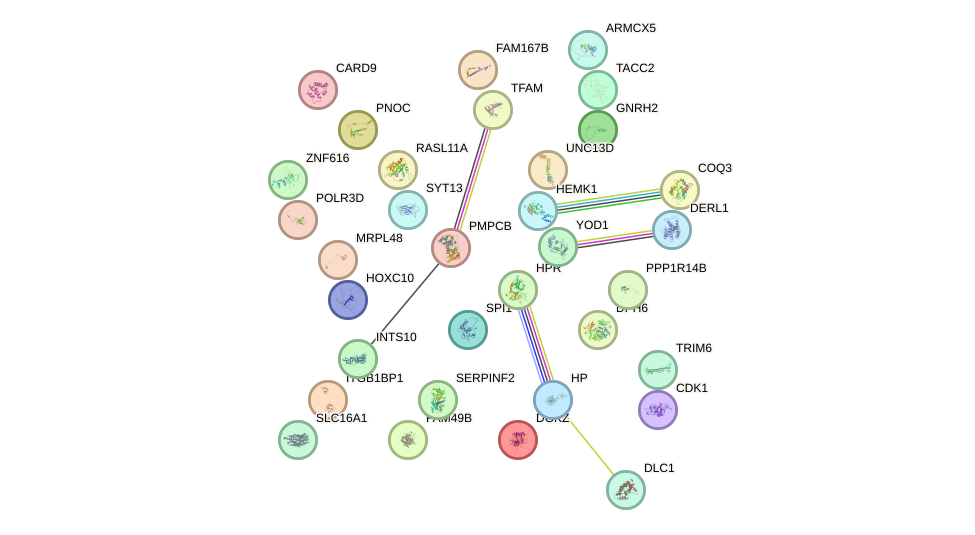
Network of associatons between targets according to the STRING database.
First level regulatory network of THRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.3 | 1.8 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.4 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.3 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 1.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.4 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |