Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for VSX1
Z-value: 0.81
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VSX1
|
ENSG00000100987.10 | visual system homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | -0.36 | 6.4e-01 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_4717835 | 0.61 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr2_-_191878874 | 0.55 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr11_-_33913708 | 0.52 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr4_-_89205705 | 0.49 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr19_-_54663473 | 0.42 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr18_-_59415987 | 0.39 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr11_+_67798090 | 0.38 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr11_-_133826852 | 0.37 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr12_+_110011571 | 0.36 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr11_+_67798114 | 0.35 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr15_+_80351910 | 0.35 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_+_159750776 | 0.33 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr6_-_32145861 | 0.32 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr17_-_1532106 | 0.31 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr6_-_30684898 | 0.31 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr20_-_17662705 | 0.29 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr7_+_100136811 | 0.29 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr20_-_17662878 | 0.28 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chrM_+_10758 | 0.28 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_+_133930798 | 0.28 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr18_-_61329118 | 0.28 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr19_+_4007644 | 0.27 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr19_+_46498704 | 0.27 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr18_+_21594384 | 0.26 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr11_-_59383617 | 0.26 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chrX_-_18690210 | 0.26 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr14_+_32798462 | 0.26 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_31783291 | 0.25 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr17_+_62223320 | 0.25 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr16_-_2264779 | 0.25 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr19_-_54618650 | 0.24 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chrM_+_4431 | 0.24 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_+_44115814 | 0.23 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr1_+_6845384 | 0.23 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_+_7694623 | 0.23 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr19_-_15529790 | 0.23 |
ENST00000596195.1
ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L
|
A kinase (PRKA) anchor protein 8-like |
| chr19_-_54619006 | 0.22 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr2_-_240322643 | 0.22 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr14_-_24584138 | 0.22 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr19_-_6737576 | 0.22 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chr4_+_169013666 | 0.22 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_+_186288454 | 0.22 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr1_+_154947126 | 0.22 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr2_+_149402989 | 0.21 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_-_25451065 | 0.21 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr19_+_19303720 | 0.20 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr8_-_131399110 | 0.20 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr17_+_79650962 | 0.20 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr1_-_36916066 | 0.20 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr11_-_95523500 | 0.20 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr2_-_70780770 | 0.20 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr2_-_70520832 | 0.20 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr19_-_7694417 | 0.19 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr1_-_154946792 | 0.19 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_+_4153598 | 0.19 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr6_-_26124138 | 0.19 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr19_+_1103936 | 0.19 |
ENST00000354171.8
ENST00000589115.1 |
GPX4
|
glutathione peroxidase 4 |
| chr19_+_56652686 | 0.18 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr6_-_26271612 | 0.18 |
ENST00000305910.3
|
HIST1H3G
|
histone cluster 1, H3g |
| chr1_+_154947148 | 0.18 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr19_-_40324767 | 0.18 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_+_50180409 | 0.18 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_-_121986923 | 0.18 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr19_+_17420340 | 0.18 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr11_+_67798363 | 0.18 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_+_16222439 | 0.17 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr16_-_67965756 | 0.17 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr19_+_19303572 | 0.17 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chrX_+_19373700 | 0.17 |
ENST00000379804.1
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr19_+_50270219 | 0.17 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr19_+_54641444 | 0.17 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr6_-_27860956 | 0.17 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chrX_-_77150911 | 0.17 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr6_-_31782813 | 0.17 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr4_-_89205879 | 0.17 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr6_-_62996066 | 0.17 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr1_-_23520755 | 0.16 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr1_+_209602156 | 0.16 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr19_-_46195029 | 0.16 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr19_+_39421556 | 0.16 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chrX_+_151999511 | 0.15 |
ENST00000370274.3
ENST00000440023.1 ENST00000432467.1 |
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like |
| chr3_-_42003479 | 0.15 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr18_-_71959159 | 0.15 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr17_+_79651007 | 0.15 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr17_-_74722536 | 0.15 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr9_+_1050331 | 0.15 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr16_-_4784128 | 0.15 |
ENST00000592711.1
ENST00000590147.1 ENST00000304283.4 ENST00000592190.1 ENST00000589065.1 ENST00000585773.1 ENST00000450067.2 ENST00000592698.1 ENST00000586166.1 ENST00000586605.1 ENST00000592421.1 |
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr9_-_38424443 | 0.15 |
ENST00000377694.1
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1 |
| chr14_-_74959978 | 0.15 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chrX_+_46696372 | 0.15 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr17_+_46970127 | 0.14 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_-_150138246 | 0.14 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr1_+_151138526 | 0.14 |
ENST00000368902.1
|
SCNM1
|
sodium channel modifier 1 |
| chr4_-_87515202 | 0.14 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_+_22501658 | 0.14 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr17_-_74722672 | 0.14 |
ENST00000397625.4
ENST00000445478.2 |
JMJD6
|
jumonji domain containing 6 |
| chr16_+_3184924 | 0.14 |
ENST00000574902.1
ENST00000396878.3 |
ZNF213
|
zinc finger protein 213 |
| chr13_-_50018241 | 0.14 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr19_+_18726786 | 0.14 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr7_+_99699179 | 0.14 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr19_+_10828724 | 0.13 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr17_+_46970178 | 0.13 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr15_-_41522889 | 0.13 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr19_+_10828795 | 0.13 |
ENST00000389253.4
ENST00000355667.6 ENST00000408974.4 |
DNM2
|
dynamin 2 |
| chr1_-_154946825 | 0.13 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr5_+_177631497 | 0.13 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr16_+_69458537 | 0.13 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr19_+_54619125 | 0.13 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr6_-_94129244 | 0.13 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr19_-_6424783 | 0.13 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr16_+_68119440 | 0.13 |
ENST00000346183.3
ENST00000329524.4 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr3_+_47324424 | 0.13 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr19_-_19302931 | 0.13 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr7_-_122339162 | 0.13 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr14_+_29236269 | 0.13 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr13_-_50018140 | 0.13 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr4_-_82393052 | 0.13 |
ENST00000335927.7
ENST00000504863.1 ENST00000264400.2 |
RASGEF1B
|
RasGEF domain family, member 1B |
| chr16_+_23652773 | 0.12 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr13_-_41240717 | 0.12 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr16_-_30773372 | 0.12 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr20_+_36322408 | 0.12 |
ENST00000361383.6
ENST00000447935.1 ENST00000405275.2 |
CTNNBL1
|
catenin, beta like 1 |
| chr11_-_66056596 | 0.12 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr7_-_99699538 | 0.12 |
ENST00000343023.6
ENST00000303887.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_-_150669604 | 0.12 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr19_-_14228541 | 0.12 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr1_-_78444738 | 0.12 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr19_-_40324255 | 0.12 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr6_+_31795506 | 0.12 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr1_-_149900122 | 0.12 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr17_-_74733404 | 0.12 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr20_+_16729003 | 0.12 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr15_+_73735490 | 0.12 |
ENST00000331090.6
ENST00000560581.1 |
C15orf60
|
chromosome 15 open reading frame 60 |
| chrX_-_153363125 | 0.12 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr15_+_83478370 | 0.12 |
ENST00000286760.4
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr16_+_57844549 | 0.12 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr19_+_1205740 | 0.11 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr11_+_63754294 | 0.11 |
ENST00000543988.1
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr17_-_66453562 | 0.11 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr8_-_29120604 | 0.11 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr7_+_99933730 | 0.11 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr12_+_7282795 | 0.11 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr22_+_39916558 | 0.11 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr17_+_79981144 | 0.11 |
ENST00000306688.3
|
LRRC45
|
leucine rich repeat containing 45 |
| chr1_-_47655686 | 0.11 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr14_-_24711470 | 0.11 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr17_-_42277203 | 0.11 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr19_-_39421377 | 0.11 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr6_-_49755019 | 0.11 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr19_-_39881669 | 0.11 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr7_+_916183 | 0.11 |
ENST00000265857.3
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr17_-_47785265 | 0.11 |
ENST00000511763.1
ENST00000515850.1 ENST00000415270.2 ENST00000240333.6 |
SLC35B1
|
solute carrier family 35, member B1 |
| chr9_+_34646624 | 0.11 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr1_-_78444776 | 0.11 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr10_-_50970322 | 0.11 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr10_-_21186144 | 0.11 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr1_+_43148059 | 0.11 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr1_-_190446759 | 0.10 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr5_+_177631523 | 0.10 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr3_+_130279178 | 0.10 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr6_-_66417107 | 0.10 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr11_-_57479673 | 0.10 |
ENST00000337672.2
ENST00000431606.2 |
MED19
|
mediator complex subunit 19 |
| chr22_-_30960876 | 0.10 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_+_36195429 | 0.10 |
ENST00000392197.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr6_-_33385902 | 0.10 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr7_-_99716952 | 0.10 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr13_+_19756173 | 0.10 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr11_+_65554493 | 0.10 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr10_-_99205607 | 0.10 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr19_-_14606900 | 0.10 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr19_+_50180507 | 0.10 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_+_15699286 | 0.10 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr14_+_32798547 | 0.10 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_+_179125907 | 0.10 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr15_+_80351977 | 0.10 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr11_+_63753883 | 0.10 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr17_-_79980734 | 0.10 |
ENST00000584600.1
ENST00000584347.1 ENST00000580435.1 ENST00000306704.6 ENST00000392359.3 |
STRA13
|
stimulated by retinoic acid 13 |
| chr2_-_69664586 | 0.10 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr1_+_92414928 | 0.10 |
ENST00000362005.3
ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT
|
bromodomain, testis-specific |
| chr2_-_217560248 | 0.09 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr19_-_6737241 | 0.09 |
ENST00000430424.4
ENST00000597298.1 |
GPR108
|
G protein-coupled receptor 108 |
| chr16_+_69458428 | 0.09 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr10_-_131909071 | 0.09 |
ENST00000456581.1
|
LINC00959
|
long intergenic non-protein coding RNA 959 |
| chr19_-_47616992 | 0.09 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr1_-_45956868 | 0.09 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr14_-_71276211 | 0.09 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr12_+_53773944 | 0.09 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr16_-_58663720 | 0.09 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr3_-_105588231 | 0.09 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr14_-_24711806 | 0.09 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr16_+_84224780 | 0.09 |
ENST00000315906.5
|
ADAD2
|
adenosine deaminase domain containing 2 |
| chr6_-_33385870 | 0.09 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr22_+_17956618 | 0.09 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr5_+_33440802 | 0.09 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr17_+_46970134 | 0.09 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr7_-_99717463 | 0.09 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_47841485 | 0.09 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr19_+_41770269 | 0.09 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr22_-_42486747 | 0.09 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr18_+_3411595 | 0.09 |
ENST00000552383.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_-_119759795 | 0.09 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
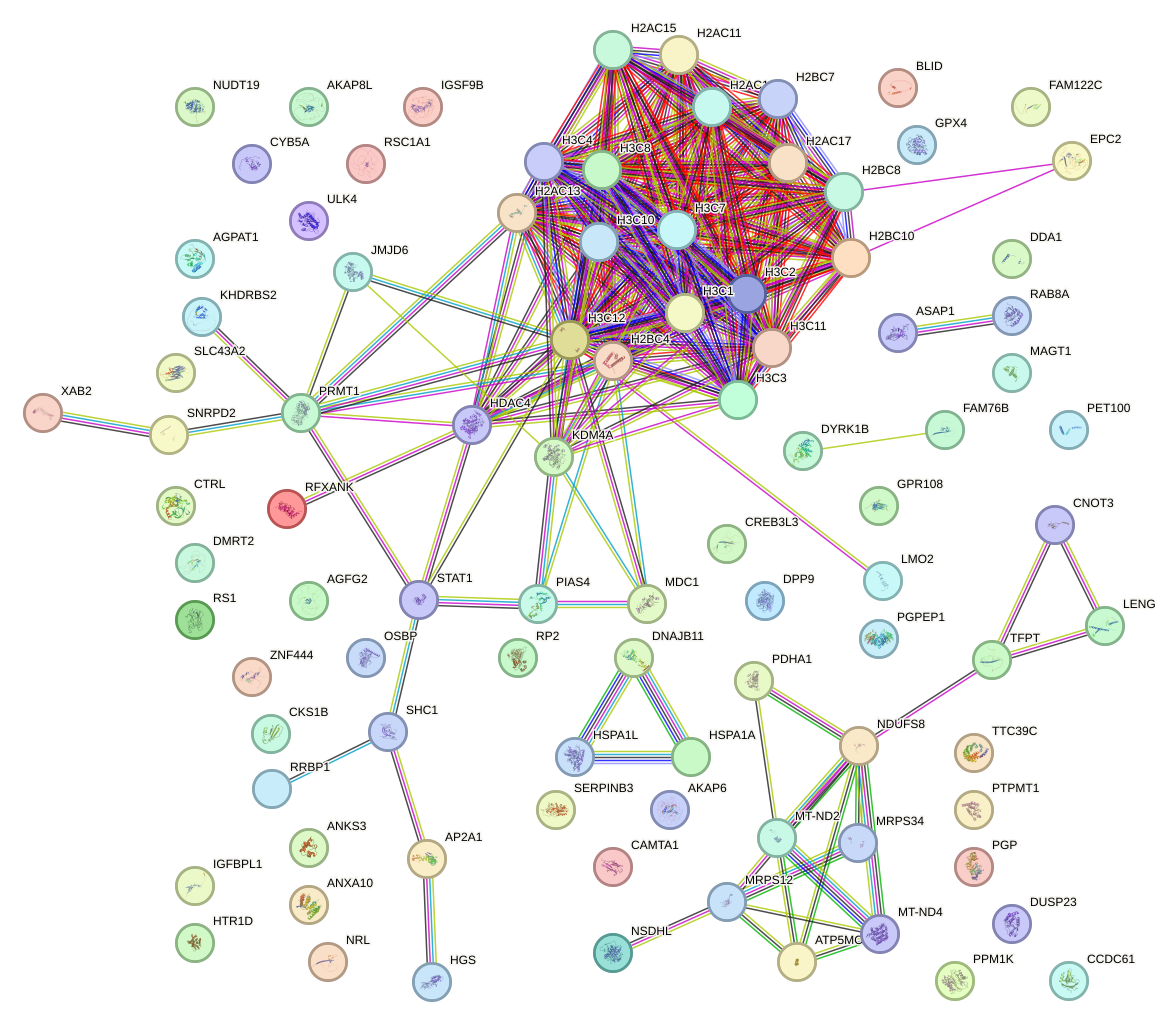
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.3 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.2 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.4 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0060770 | positive thymic T cell selection(GO:0045059) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 1.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |