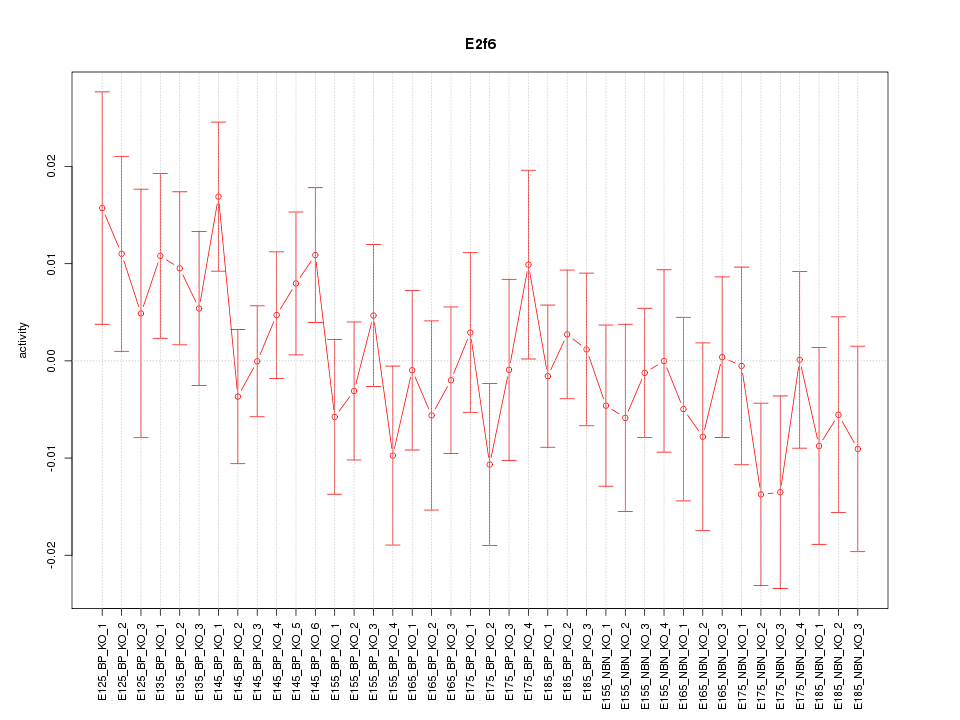
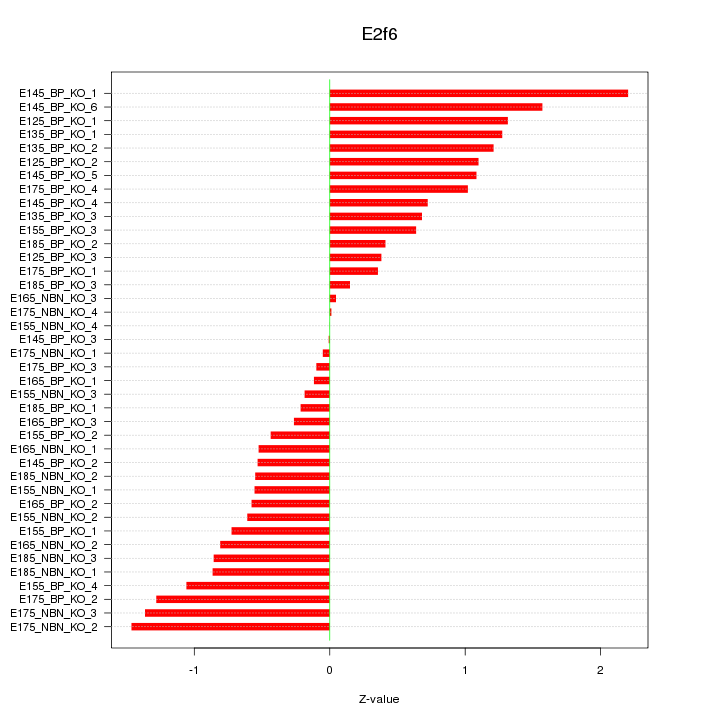
Motif ID: E2f6
Z-value: 0.853

Transcription factors associated with E2f6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f6 | ENSMUSG00000057469.7 | E2f6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f6 | mm10_v2_chr12_+_16810940_16810978 | 0.72 | 2.1e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.3 | 4.0 | GO:0030421 | defecation(GO:0030421) |
| 0.8 | 2.5 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.8 | 3.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.7 | 0.7 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.7 | 2.7 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.6 | 1.3 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.6 | 3.2 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.6 | 1.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 | 1.8 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.6 | 2.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 0.6 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.6 | 1.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.6 | 2.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.6 | 1.1 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.5 | 6.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.5 | 2.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.5 | 5.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.5 | 2.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.5 | 5.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.5 | 1.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.5 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 1.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.5 | 1.4 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 3.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.5 | 1.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.5 | 2.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.5 | 2.3 | GO:0007223 | muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.5 | 1.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 1.8 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 | 1.3 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.4 | 0.9 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.4 | 0.8 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.4 | 2.5 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.4 | 2.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.4 | 2.7 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.4 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.4 | 1.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.4 | 1.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.4 | 1.1 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.3 | 1.4 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 1.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 3.0 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.3 | 5.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 0.7 | GO:0072180 | mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.3 | 1.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.0 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.3 | 0.9 | GO:1904978 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.3 | 2.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 1.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.3 | 5.5 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.3 | 1.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 0.9 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.3 | 1.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.3 | 1.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 | 3.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.3 | 1.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 1.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.0 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.0 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 1.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 | 3.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 | 1.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 1.1 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.2 | 0.2 | GO:1901563 | response to camptothecin(GO:1901563) |
| 0.2 | 1.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 3.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 4.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 0.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.9 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.2 | 1.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 1.4 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 1.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.2 | 1.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 1.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 0.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 2.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 1.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.7 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 1.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 1.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 0.7 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.2 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.5 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 0.5 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.2 | 0.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 0.5 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.2 | 1.9 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.2 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 0.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.2 | 0.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 0.6 | GO:0035021 | amygdala development(GO:0021764) negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 1.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 0.6 | GO:2001045 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.6 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.5 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 1.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 4.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.0 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 1.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.0 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 0.4 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.1 | 1.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.7 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.7 | GO:0071372 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 1.5 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.1 | 0.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.7 | GO:1903317 | regulation of protein processing(GO:0070613) regulation of protein maturation(GO:1903317) |
| 0.1 | 0.9 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.7 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.9 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.1 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.8 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 1.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 0.3 | GO:0060558 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.1 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.4 | GO:0045654 | thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.8 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:2000323 | response to redox state(GO:0051775) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 2.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 1.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.9 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.9 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.1 | 3.0 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.7 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.5 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 1.0 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.7 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.1 | 2.2 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 1.8 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.6 | GO:0070423 | response to peptidoglycan(GO:0032494) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 2.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 2.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.2 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) negative regulation of metalloendopeptidase activity(GO:1904684) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 2.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.1 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 | 0.1 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 1.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.6 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.5 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.8 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.3 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:1904152 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.0 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) left lung development(GO:0060459) cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.4 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 1.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.0 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.8 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.9 | 2.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.5 | 1.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 1.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.4 | 5.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 1.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 0.9 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 1.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 2.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 1.1 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.3 | 1.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 1.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 1.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 1.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.9 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.4 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 2.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 3.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 5.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 0.3 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.1 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.9 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 5.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 8.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 4.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.0 | 5.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.9 | 5.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 2.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.7 | 2.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.6 | 2.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.6 | 2.3 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.5 | 1.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.4 | 1.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 1.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.4 | 1.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.4 | 1.1 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.3 | 2.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 2.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 2.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 0.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 2.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 1.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 1.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.3 | 2.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 1.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 1.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 6.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 5.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 1.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 0.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 0.5 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 4.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 3.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 1.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 2.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.5 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 4.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 3.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.3 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.5 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 3.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 3.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 4.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 3.8 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 1.9 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 1.9 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 13.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 6.7 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 8.3 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 6.7 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 6.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 3.8 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.1 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.9 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 1.1 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 4.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.4 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 2.9 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.8 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.3 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.1 | 1.8 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 0.4 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 1.6 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.4 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 0.6 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.5 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.1 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.6 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.8 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID_MYC_PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 6.1 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 4.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 2.1 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 4.2 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.2 | 2.7 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 1.3 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 0.8 | REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 2.2 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.9 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.6 | REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 6.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.5 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.0 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.8 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 2.7 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.6 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.2 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.3 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.3 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 1.4 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.6 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.4 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 4.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.2 | REACTOME_E2F_MEDIATED_REGULATION_OF_DNA_REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.1 | 2.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.1 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.3 | REACTOME_P75NTR_SIGNALS_VIA_NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.1 | 0.7 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.3 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 3.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.9 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 1.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.8 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME_DOWNSTREAM_SIGNALING_EVENTS_OF_B_CELL_RECEPTOR_BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |