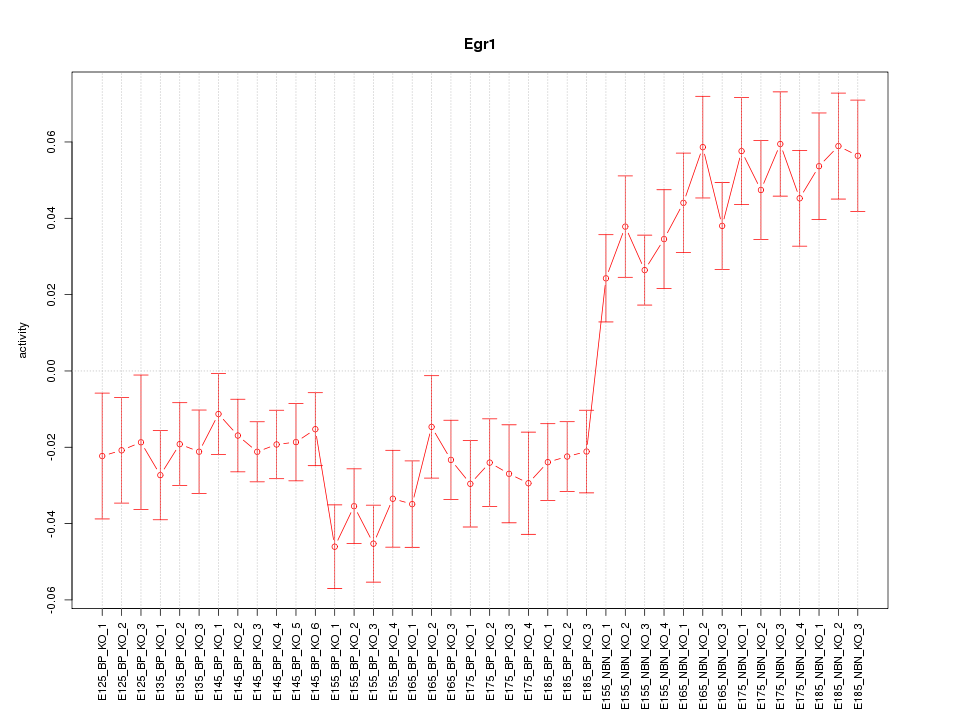
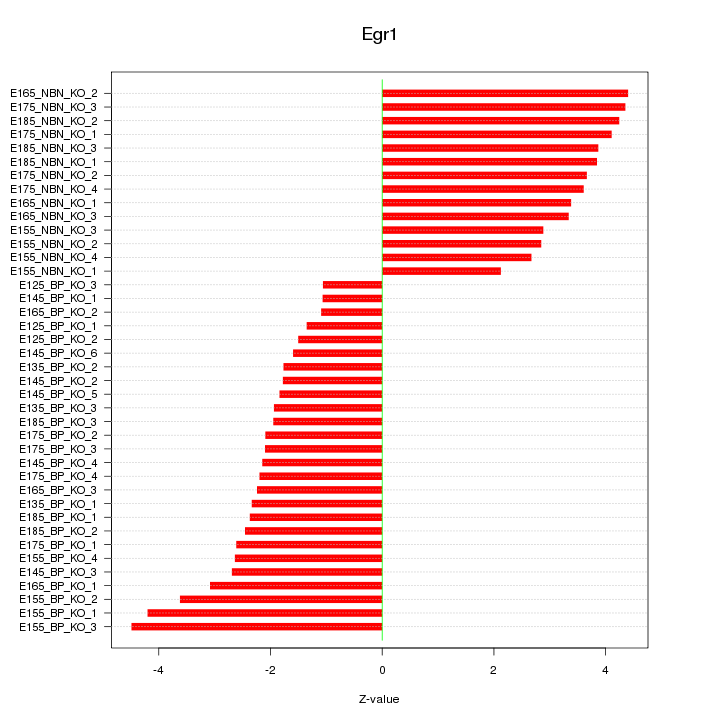
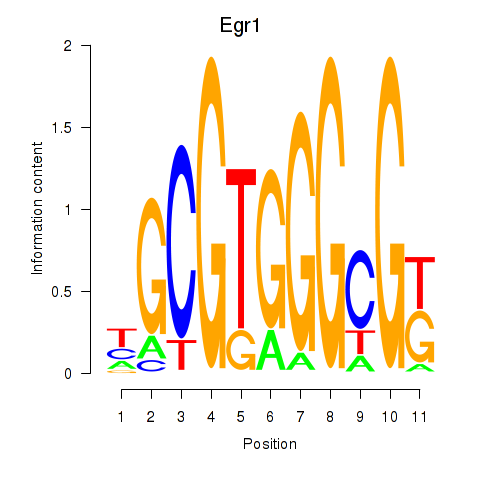
Motif ID: Egr1
Z-value: 2.869
Transcription factors associated with Egr1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr1 | ENSMUSG00000038418.7 | Egr1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm10_v2_chr18_+_34861200_34861215 | 0.10 | 5.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.9 | 47.7 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 6.1 | 18.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 4.2 | 21.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 4.0 | 12.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 3.9 | 11.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 3.4 | 10.2 | GO:0030070 | insulin processing(GO:0030070) |
| 3.2 | 15.9 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 3.1 | 9.3 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 2.9 | 8.6 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 2.9 | 14.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 2.7 | 5.4 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 2.6 | 28.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 2.3 | 27.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 2.3 | 13.8 | GO:0032796 | uropod organization(GO:0032796) |
| 2.3 | 6.8 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 2.3 | 2.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 2.2 | 8.8 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.2 | 13.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.1 | 12.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.9 | 9.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 1.9 | 5.7 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 1.9 | 5.6 | GO:0071336 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.8 | 9.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.8 | 7.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 1.8 | 5.3 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 1.7 | 3.4 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 1.7 | 8.5 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 1.7 | 6.7 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.7 | 11.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.7 | 8.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.6 | 16.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.6 | 4.9 | GO:0002925 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 1.6 | 9.4 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.6 | 3.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.5 | 7.5 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.5 | 8.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.5 | 4.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.4 | 18.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.4 | 7.2 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.4 | 5.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 1.4 | 4.3 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 1.4 | 4.1 | GO:0050883 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 1.4 | 5.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.4 | 4.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 1.3 | 20.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.3 | 23.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 1.3 | 10.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 1.3 | 6.4 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 1.3 | 2.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.3 | 5.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.3 | 5.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.2 | 25.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.2 | 4.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.2 | 6.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.2 | 13.1 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 1.2 | 20.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 1.2 | 3.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.2 | 21.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 1.2 | 4.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 1.2 | 8.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 1.1 | 5.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 1.1 | 14.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.1 | 3.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 1.0 | 4.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.0 | 2.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.0 | 4.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.0 | 6.7 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.9 | 3.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.9 | 2.7 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.9 | 2.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.9 | 8.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.9 | 2.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.9 | 5.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.9 | 5.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.8 | 10.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.8 | 4.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.8 | 19.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.8 | 8.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.7 | 4.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.7 | 2.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.7 | 2.1 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.7 | 2.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 | 5.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.6 | 3.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 4.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.6 | 4.9 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.6 | 15.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.6 | 1.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.6 | 2.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.6 | 6.8 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.6 | 7.3 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.6 | 4.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.6 | 2.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.5 | 6.4 | GO:0043084 | penile erection(GO:0043084) |
| 0.5 | 1.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 2.6 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.5 | 3.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 6.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 1.9 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.5 | 2.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.5 | 5.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.5 | 1.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 6.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.5 | 2.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.4 | 8.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.4 | 5.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.4 | 1.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 1.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 3.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.4 | 2.7 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 18.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 2.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.4 | 1.8 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 1.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 | 1.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 1.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.3 | 0.7 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 1.4 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 3.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 1.6 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.3 | 6.8 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.3 | 1.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 1.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 2.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 13.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.3 | 8.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 1.7 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 0.6 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 1.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 2.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.3 | 1.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 2.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 1.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 7.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 1.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 3.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 2.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 2.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 5.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 2.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.4 | GO:0032306 | regulation of prostaglandin secretion(GO:0032306) |
| 0.2 | 2.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 1.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 8.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 3.3 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.2 | 7.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.2 | 2.1 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 1.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 22.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 0.9 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.2 | 3.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 7.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.2 | 3.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 5.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 4.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 2.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 5.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 0.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 4.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 2.0 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 4.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.0 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 6.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.6 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.0 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 17.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 3.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.4 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.1 | 2.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.5 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 1.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 6.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 2.0 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 2.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 2.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 1.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 2.6 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 2.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.6 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 1.5 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 | 2.4 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 0.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.6 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 1.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.1 | 2.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 3.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 5.9 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 3.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 2.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.4 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 6.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 16.1 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.5 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.9 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 1.1 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 2.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.7 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:1901879 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) regulation of protein depolymerization(GO:1901879) |
| 0.0 | 0.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.4 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 1.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.4 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 1.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 18.9 | GO:0008091 | spectrin(GO:0008091) |
| 3.0 | 11.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.5 | 32.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 2.4 | 7.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 2.2 | 13.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.2 | 8.6 | GO:0090537 | CERF complex(GO:0090537) |
| 2.0 | 12.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.7 | 6.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.6 | 4.9 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 1.4 | 48.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 1.4 | 21.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.4 | 13.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.3 | 5.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.3 | 4.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.3 | 13.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.2 | 15.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.2 | 14.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.2 | 8.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.2 | 4.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.1 | 3.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.1 | 38.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.1 | 15.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.0 | 11.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.8 | 23.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.8 | 14.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.7 | 8.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.7 | 20.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 2.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 2.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.7 | 3.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 2.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 6.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.6 | 2.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 1.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.6 | 30.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.5 | 6.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 35.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 4.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 1.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.4 | 2.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 1.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.4 | 4.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 10.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 4.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 3.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 6.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 24.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 3.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 4.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 19.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.3 | 21.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 6.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 2.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 4.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 5.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 5.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 7.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 2.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 0.6 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.2 | 5.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 8.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 2.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 8.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 8.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 3.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 9.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 2.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 0.5 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 1.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 10.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 15.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 6.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 28.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 8.1 | GO:0044309 | neuron spine(GO:0044309) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 36.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 1.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 12.1 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 4.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.1 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 5.8 | 28.9 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 4.8 | 14.3 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 3.2 | 13.0 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 2.9 | 8.6 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 2.7 | 8.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.7 | 21.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 2.5 | 27.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 2.4 | 21.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 2.3 | 4.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 2.0 | 8.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 2.0 | 11.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.0 | 50.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.9 | 5.6 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.8 | 11.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.8 | 7.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.8 | 7.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.8 | 10.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.7 | 6.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.7 | 10.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.7 | 30.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.6 | 6.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 1.6 | 17.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.5 | 6.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.4 | 17.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.4 | 5.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.4 | 5.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.4 | 8.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.4 | 10.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.3 | 5.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.3 | 17.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.3 | 13.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 14.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.2 | 3.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.1 | 3.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.1 | 3.4 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 1.1 | 5.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.0 | 5.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 1.0 | 4.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 1.0 | 2.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.9 | 14.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.8 | 6.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.8 | 6.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.8 | 2.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.8 | 6.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 16.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.7 | 9.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.7 | 2.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.7 | 4.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 2.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.6 | 16.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.6 | 9.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 3.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 2.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.6 | 2.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 4.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 5.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 1.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.5 | 2.6 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.5 | 14.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.5 | 0.5 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.5 | 1.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 5.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 1.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 6.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 4.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 2.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 5.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 7.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 4.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.4 | 1.1 | GO:0019966 | C-X-C chemokine binding(GO:0019958) interleukin-1 binding(GO:0019966) tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 3.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 5.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 16.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 18.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 3.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 6.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 9.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 2.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 1.7 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 5.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 6.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 6.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 24.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 2.6 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.2 | 11.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 5.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 7.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 5.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 10.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 8.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 4.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 2.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 1.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 5.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 3.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 11.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 5.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 3.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.9 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.2 | 2.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 8.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 2.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 3.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 4.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 11.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 8.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 3.0 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 2.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 4.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 13.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 6.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 8.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 9.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 7.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 3.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 5.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 48.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.4 | 24.1 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.9 | 27.6 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.8 | 32.8 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.7 | 31.7 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.6 | 14.3 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 4.9 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 16.3 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.5 | 27.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.4 | 3.1 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.4 | 17.0 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 19.8 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.3 | 7.8 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.3 | 4.3 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.3 | 6.8 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.3 | 28.5 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 2.6 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 10.5 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 8.9 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.2 | 12.6 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.2 | 9.4 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 4.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 2.7 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 6.8 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.2 | 1.7 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.5 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 1.6 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.2 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.5 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 11.4 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.4 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 9.9 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.8 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.6 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.3 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 1.3 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 1.6 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.1 | 0.1 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.4 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.3 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.4 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 2.4 | 59.5 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.3 | 16.2 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 2.1 | 46.3 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 2.1 | 28.9 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.7 | 19.0 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 1.3 | 20.0 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.2 | 33.0 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 1.0 | 8.1 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.9 | 37.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.9 | 6.9 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.8 | 17.1 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.7 | 23.9 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.7 | 20.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.7 | 5.4 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 8.4 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.6 | 15.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.5 | 13.0 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.5 | 8.0 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 7.9 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 5.4 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.5 | 5.6 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.4 | 10.8 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 4.9 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 3.0 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.4 | 10.9 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.3 | 5.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 3.1 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.2 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 20.6 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.3 | 4.3 | REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 5.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.1 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.8 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.8 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 2.8 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 9.1 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 5.7 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 1.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 3.1 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 12.2 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.2 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.6 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.1 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.1 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 5.0 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 7.7 | REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 1.2 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.1 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.6 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.0 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.5 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 3.2 | REACTOME_TCR_SIGNALING | Genes involved in TCR signaling |
| 0.1 | 4.8 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 3.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.6 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 6.7 | REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 3.9 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.1 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.6 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.4 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |