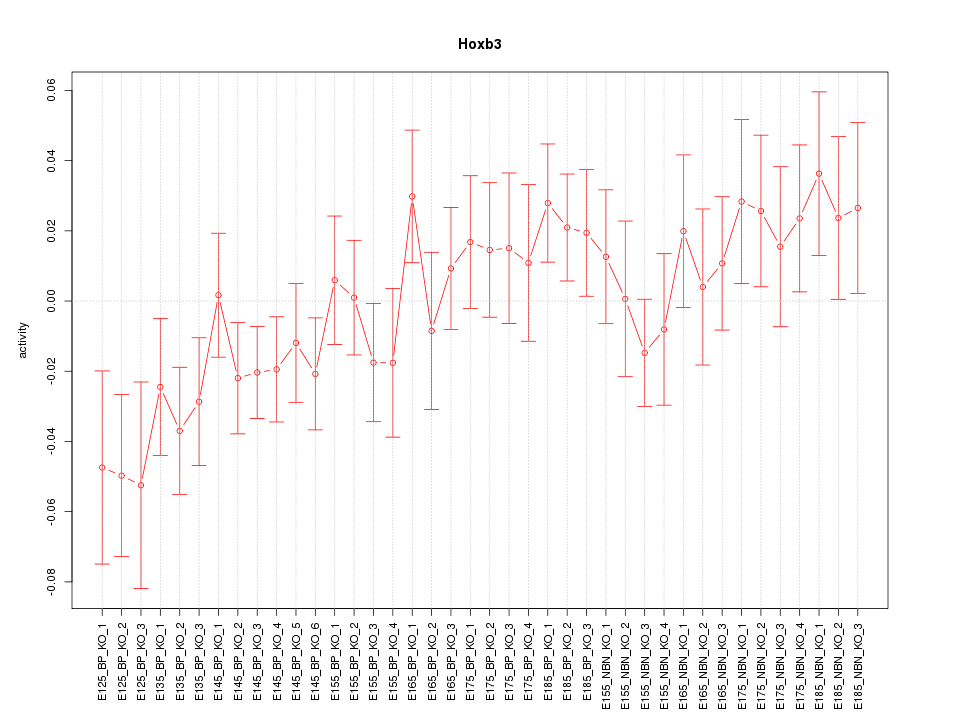
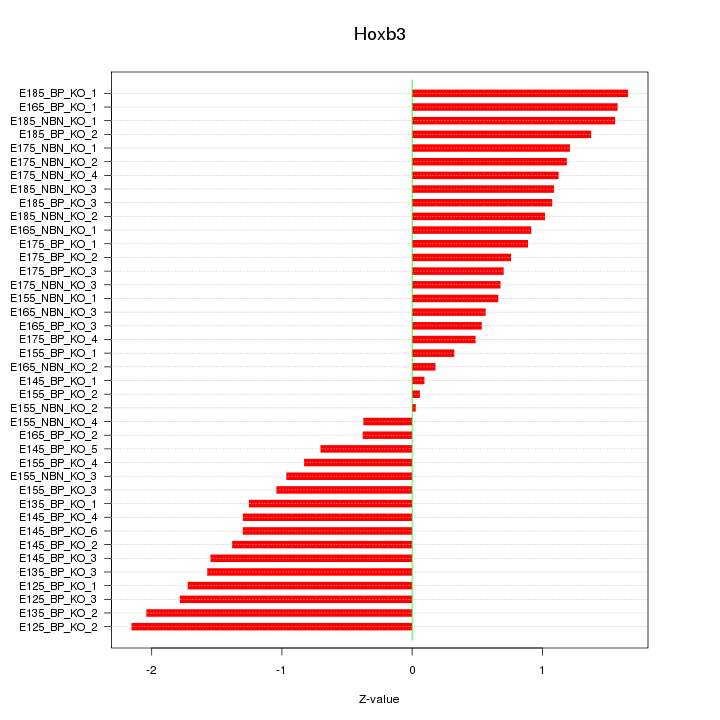
Motif ID: Hoxb3
Z-value: 1.139
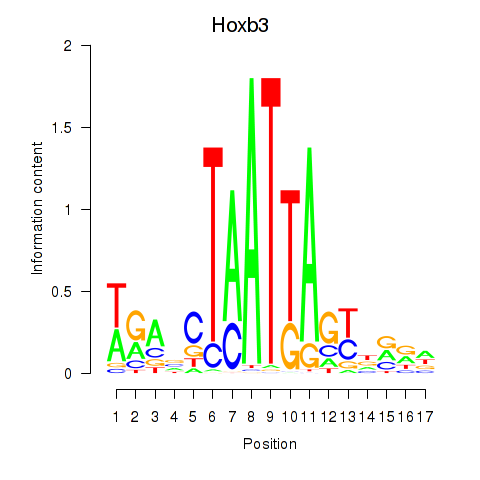
Transcription factors associated with Hoxb3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb3 | ENSMUSG00000048763.5 | Hoxb3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.1 | 15.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.3 | 10.6 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 1.3 | 6.3 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 1.2 | 5.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 1.2 | 4.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.1 | 4.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.9 | 3.7 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.9 | 2.7 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.7 | 2.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 3.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 1.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 2.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.3 | 1.8 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 7.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 3.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 2.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 1.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 5.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 2.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 2.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 1.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 4.1 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 3.4 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 6.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 3.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 33.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 11.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 9.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.6 | 10.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.5 | 4.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.4 | 15.0 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 1.3 | 6.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.1 | 4.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 5.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 1.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.3 | 1.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 3.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 0.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 2.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 4.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 2.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 4.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 5.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 20.6 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 15.6 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 6.3 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.3 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.0 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.4 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.3 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.5 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.7 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |