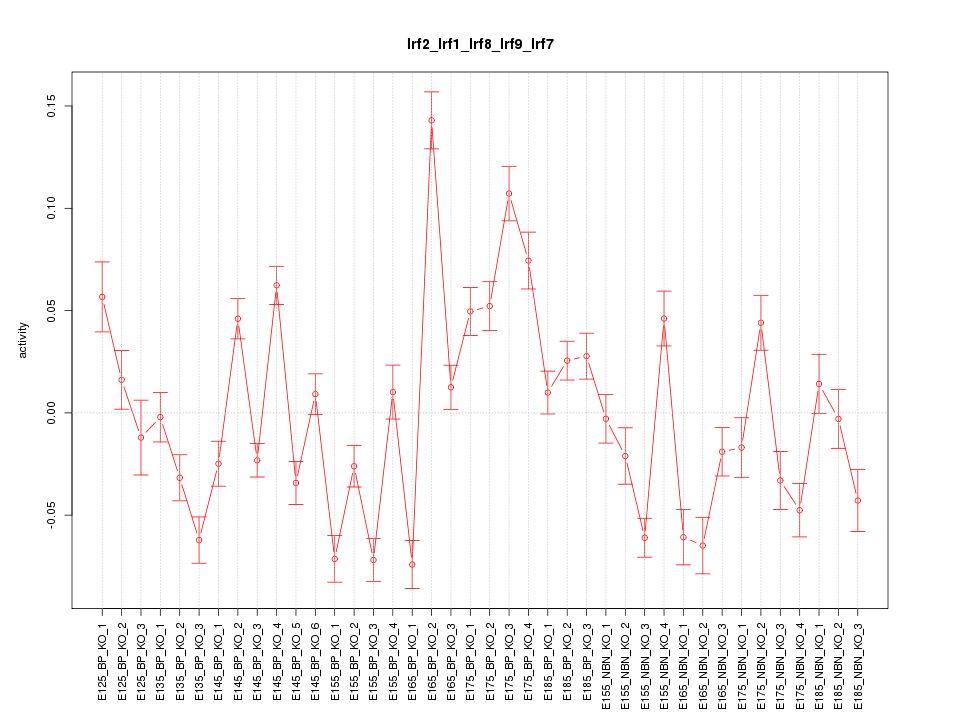
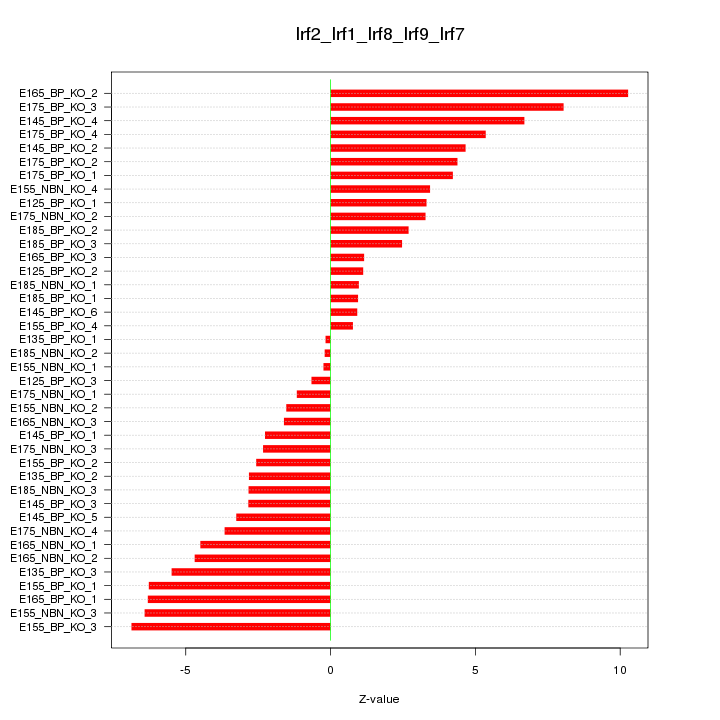
Motif ID: Irf2_Irf1_Irf8_Irf9_Irf7
Z-value: 4.085
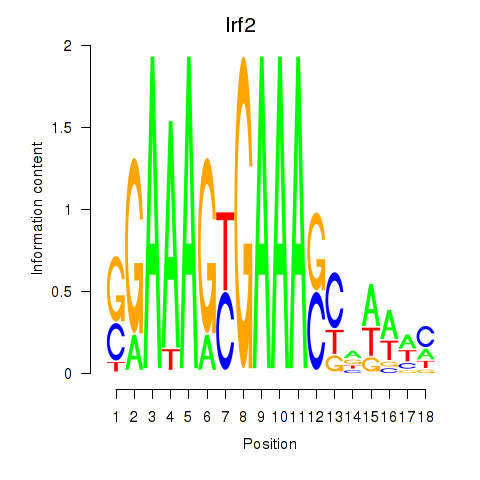
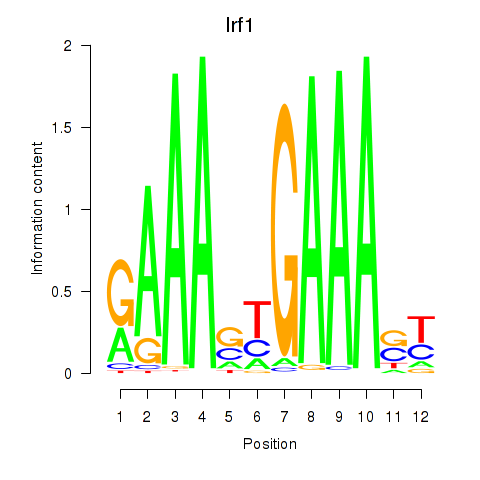
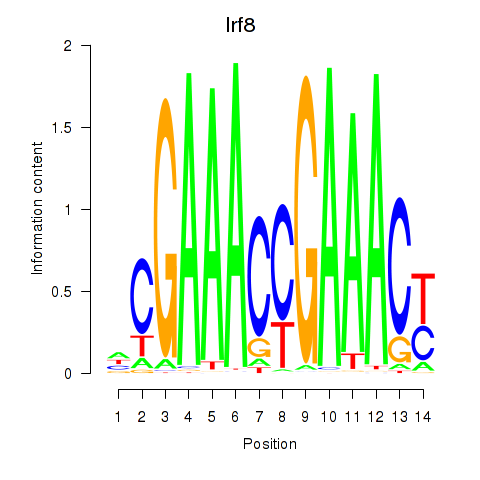
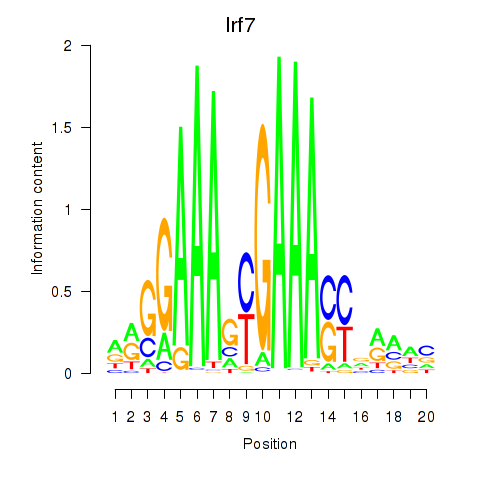
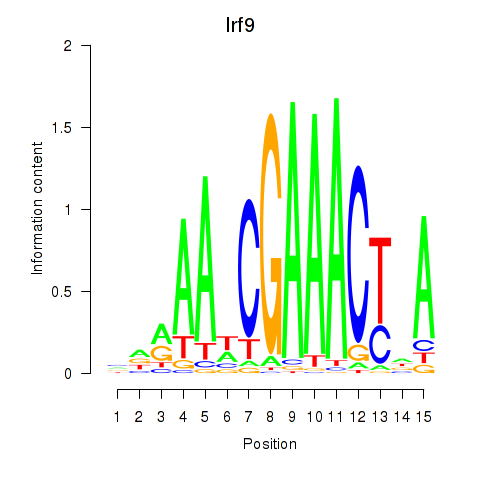
Transcription factors associated with Irf2_Irf1_Irf8_Irf9_Irf7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Irf1 | ENSMUSG00000018899.10 | Irf1 |
| Irf2 | ENSMUSG00000031627.7 | Irf2 |
| Irf7 | ENSMUSG00000025498.8 | Irf7 |
| Irf8 | ENSMUSG00000041515.3 | Irf8 |
| Irf9 | ENSMUSG00000002325.8 | Irf9 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf1 | mm10_v2_chr11_+_53770014_53770057 | 0.59 | 5.9e-05 | Click! |
| Irf7 | mm10_v2_chr7_-_141266415_141266481 | 0.53 | 4.7e-04 | Click! |
| Irf8 | mm10_v2_chr8_+_120736352_120736385 | 0.39 | 1.3e-02 | Click! |
| Irf2 | mm10_v2_chr8_+_46739745_46739791 | 0.30 | 6.3e-02 | Click! |
| Irf9 | mm10_v2_chr14_+_55604550_55604579 | 0.29 | 7.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 37.7 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 6.2 | 24.9 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 5.5 | 110.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 5.5 | 27.4 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 5.2 | 15.6 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 4.9 | 14.6 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 4.0 | 12.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.7 | 11.2 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 3.5 | 28.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 3.1 | 9.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 2.9 | 25.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 2.8 | 96.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 2.5 | 9.9 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 2.3 | 13.5 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 2.2 | 6.7 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 2.1 | 12.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 1.9 | 7.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.8 | 1.8 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 1.8 | 5.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.8 | 3.6 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 1.8 | 12.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 1.5 | 6.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.5 | 12.2 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 1.5 | 1.5 | GO:2000564 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.4 | 6.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.4 | 6.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.2 | 7.4 | GO:1900225 | NLRP3 inflammasome complex assembly(GO:0044546) regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 1.2 | 2.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 1.2 | 4.8 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 1.2 | 13.0 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.2 | 3.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.0 | 3.1 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 1.0 | 3.0 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) |
| 0.9 | 3.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.9 | 1.8 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.8 | 21.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.8 | 11.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.8 | 2.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.7 | 5.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.7 | 2.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 5.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.6 | 1.9 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 3.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.6 | 1.8 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 1.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.6 | 1.8 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.6 | 2.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 2.7 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.5 | 2.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.5 | 2.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 3.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.5 | 2.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.5 | 1.4 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.5 | 2.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 15.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.4 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 3.0 | GO:0044838 | cell quiescence(GO:0044838) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.4 | 1.7 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 | 13.5 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.4 | 1.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 1.6 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 3.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 1.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 2.6 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.4 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.0 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.3 | 1.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.3 | 0.3 | GO:0071649 | chemokine (C-C motif) ligand 5 production(GO:0071609) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.3 | 1.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 1.6 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 0.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 1.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.3 | 2.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 0.9 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.3 | 4.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 0.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 2.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 1.4 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.3 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.3 | 3.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 0.3 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.3 | 2.8 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.3 | 2.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.3 | 1.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 1.0 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 | 1.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 0.2 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.2 | 2.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 0.7 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.2 | 1.4 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.2 | 10.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 2.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.2 | 4.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.2 | 10.0 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 0.2 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.2 | 1.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.6 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.2 | 0.8 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.2 | 0.6 | GO:0070172 | oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 3.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 2.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.5 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 0.7 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 4.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 0.5 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 0.3 | GO:0002865 | immune response-inhibiting signal transduction(GO:0002765) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.2 | 2.3 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.2 | 0.2 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.2 | 0.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 4.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.7 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 0.4 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.6 | GO:0097211 | prolactin secretion(GO:0070459) response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 3.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.6 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.6 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 2.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 6.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.4 | GO:1902031 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.6 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.3 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 1.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 2.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 0.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 1.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.4 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 8.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.3 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 0.3 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 2.5 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.4 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 1.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 1.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0030222 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil differentiation(GO:0030222) primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.8 | GO:0001963 | synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 | 0.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 0.2 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.5 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.0 | 0.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 2.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.3 | GO:0043921 | modulation by host of viral transcription(GO:0043921) modulation of transcription in other organism involved in symbiotic interaction(GO:0052312) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:2000561 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:2000338 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 2.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.2 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.0 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.0 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 108.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 4.2 | 50.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 2.2 | 6.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 2.2 | 15.1 | GO:0042825 | TAP complex(GO:0042825) |
| 2.0 | 10.1 | GO:1990462 | omegasome(GO:1990462) |
| 1.8 | 27.3 | GO:0044754 | autolysosome(GO:0044754) |
| 1.6 | 6.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.1 | 8.8 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.8 | 13.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 6.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 3.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 2.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.6 | 3.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.6 | 5.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.6 | 1.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 3.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 2.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.4 | 1.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 3.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 5.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 4.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 1.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.3 | 1.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 1.0 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 2.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 42.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 7.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 0.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 15.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 1.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 5.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 9.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 4.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 14.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 507.1 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 17.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 3.4 | 13.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 2.8 | 11.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 2.7 | 8.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.5 | 27.7 | GO:0046977 | TAP binding(GO:0046977) |
| 2.4 | 14.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 2.4 | 12.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.4 | 7.2 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 2.0 | 17.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 1.9 | 7.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.8 | 9.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.7 | 6.9 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.6 | 6.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.4 | 17.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.3 | 17.4 | GO:0031386 | protein tag(GO:0031386) |
| 1.3 | 6.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 3.4 | GO:0019002 | GMP binding(GO:0019002) |
| 1.0 | 29.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.0 | 3.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.9 | 2.8 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.8 | 2.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.8 | 5.5 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.8 | 3.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 2.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.7 | 13.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.7 | 2.0 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.6 | 4.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.6 | 1.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.6 | 13.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.6 | 7.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.6 | 2.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.6 | 2.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.5 | 138.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.5 | 1.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 1.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 3.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.5 | 1.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 2.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 3.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 6.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 2.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.4 | 25.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 7.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 1.0 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 2.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 1.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 1.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 6.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 2.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 1.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 5.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 45.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.7 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.2 | 5.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.2 | 9.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 0.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.5 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 0.5 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.2 | 1.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 2.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 4.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 4.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 5.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 6.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 4.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 5.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 6.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 6.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 5.8 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 7.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 241.6 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.9 | 50.6 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.6 | 26.3 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 0.4 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 2.6 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 5.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.2 | 20.6 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 12.0 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 7.0 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.2 | 5.4 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.2 | 3.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.2 | 12.0 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 3.0 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 3.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 4.5 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.9 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 6.0 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 12.8 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 7.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 1.7 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.6 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.4 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.3 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.5 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.2 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 11.9 | NABA_CORE_MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 1.2 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.3 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.8 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 1.4 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 0.2 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 0.4 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 1.6 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.6 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 2.4 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.4 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.0 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 5.7 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.0 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.3 | 103.2 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 1.8 | 44.1 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 1.0 | 3.8 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 10.7 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.8 | 6.2 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.7 | 13.0 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.7 | 8.4 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 8.3 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.6 | 6.5 | REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.4 | 9.6 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 2.5 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 9.1 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 6.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 6.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 3.8 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 2.8 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.2 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 0.2 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.9 | REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 4.2 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.0 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 5.0 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.6 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 5.5 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 3.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.5 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.5 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.6 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.8 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 6.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.6 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 6.2 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.7 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.0 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 0.8 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.7 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 0.9 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.5 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.5 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.5 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.7 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.4 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 2.9 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME_CD28_CO_STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.8 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.1 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.3 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.1 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |