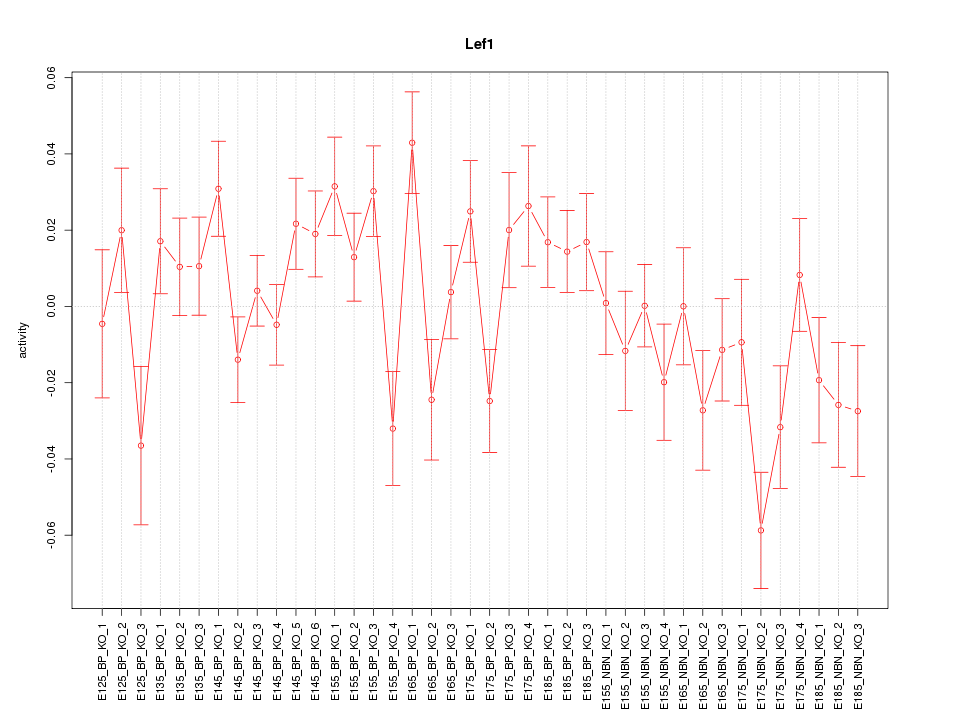
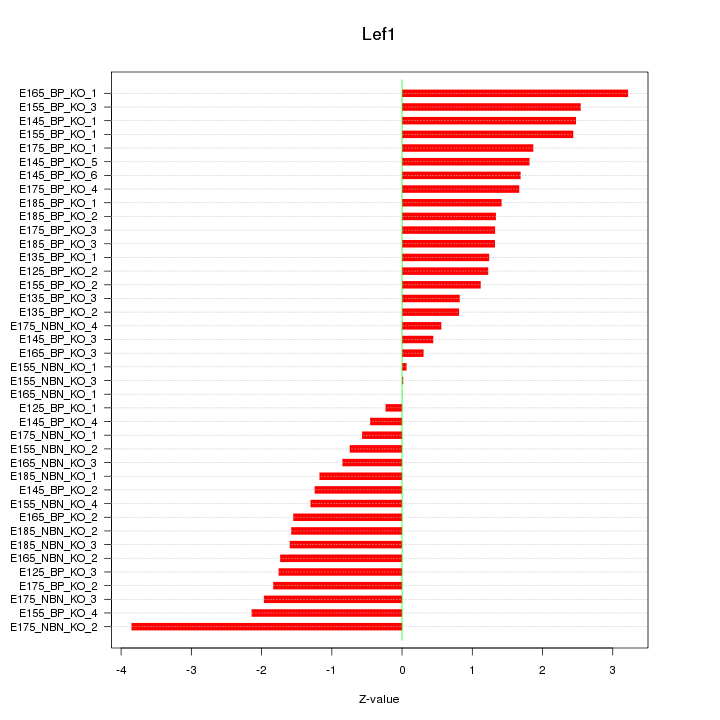
Motif ID: Lef1
Z-value: 1.594
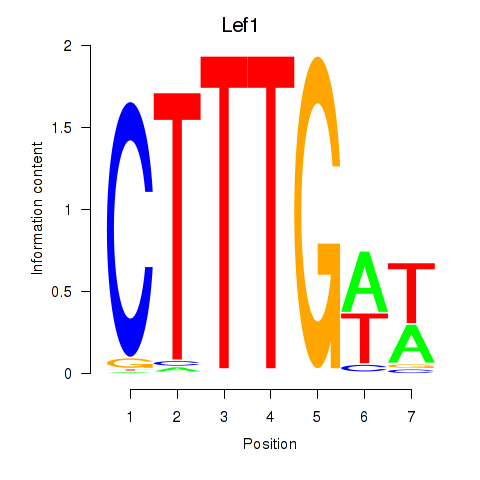
Transcription factors associated with Lef1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lef1 | ENSMUSG00000027985.8 | Lef1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | mm10_v2_chr3_+_131110350_131110471 | 0.59 | 5.9e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 2.9 | 8.7 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 2.1 | 6.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 2.1 | 6.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 1.9 | 1.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.8 | 8.9 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.5 | 4.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.4 | 4.1 | GO:0060023 | soft palate development(GO:0060023) |
| 1.3 | 21.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.3 | 5.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.2 | 6.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.2 | 5.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.2 | 4.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.1 | 3.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.1 | 3.4 | GO:1902256 | endocardial cushion fusion(GO:0003274) apoptotic process involved in outflow tract morphogenesis(GO:0003275) atrial septum primum morphogenesis(GO:0003289) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 1.1 | 7.9 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.1 | 11.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.0 | 3.1 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.0 | 5.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.0 | 3.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.9 | 7.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.8 | 5.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.8 | 5.8 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.8 | 5.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.8 | 2.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.8 | 8.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.7 | 3.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.7 | 3.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 7.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.6 | 2.5 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.6 | 1.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.6 | 1.8 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.5 | 2.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.5 | 2.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.5 | 0.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.5 | 1.5 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.5 | 1.5 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.5 | 8.6 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.5 | 8.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.4 | 1.3 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) axial mesoderm morphogenesis(GO:0048319) |
| 0.4 | 3.5 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.4 | 2.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 2.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 4.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.4 | 2.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.4 | 1.2 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.4 | 1.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 | 2.7 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.4 | 2.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.4 | 1.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.4 | 1.8 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 1.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 5.3 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.3 | 1.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 3.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 2.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 2.6 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.3 | 6.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.3 | 1.9 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 | 7.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.3 | 1.8 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.3 | 0.5 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 2.9 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.2 | 1.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 3.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.2 | 0.7 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 5.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 | 3.9 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 3.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 1.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 2.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 3.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.2 | 9.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 1.3 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 2.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.5 | GO:1990927 | negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 1.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 4.9 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 1.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 0.8 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 2.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 1.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.7 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.1 | 1.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 2.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.4 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 2.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 2.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 2.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 1.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 8.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 7.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.8 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 2.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.8 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 1.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 1.3 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 2.5 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.8 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 3.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 2.1 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 2.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 1.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 6.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 2.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 1.3 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 2.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.8 | GO:0008623 | CHRAC(GO:0008623) |
| 1.9 | 7.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.4 | 4.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.1 | 8.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.1 | 11.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.6 | 3.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.3 | 1.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 1.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 3.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 1.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 3.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 6.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 5.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 4.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.2 | 1.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 3.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 5.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 1.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 3.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 2.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 8.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 6.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 2.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 31.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 4.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 4.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 11.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.3 | 10.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.0 | 6.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.0 | 14.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 3.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.8 | 3.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 4.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.8 | 4.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.6 | 11.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 1.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.5 | 1.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 3.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 3.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.4 | 8.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 1.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 1.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 6.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 1.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 2.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 2.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 7.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.3 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 2.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 0.8 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.3 | 4.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 3.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 4.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 2.8 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 1.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 1.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 10.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 7.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 3.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.7 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 1.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 3.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 5.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 22.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 4.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 9.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 4.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 3.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 4.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 50.3 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.1 | 1.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 3.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 4.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 3.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.4 | GO:0000975 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 1.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 9.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 6.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.7 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 21.5 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.4 | 12.0 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.3 | 28.9 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 6.5 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 2.1 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 12.0 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 8.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 4.9 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.5 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.6 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.1 | 2.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 7.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.6 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 1.6 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.3 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.7 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | ST_STAT3_PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.5 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.6 | 10.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.4 | 13.0 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 5.7 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 10.6 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.2 | 6.2 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.5 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 0.8 | REACTOME_SIGNALING_BY_FGFR_IN_DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.2 | 3.1 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.7 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.7 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 4.2 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.1 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 8.9 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 4.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.6 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.1 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.5 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 3.5 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.1 | 5.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.7 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.8 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.5 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME_RAF_MAP_KINASE_CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |