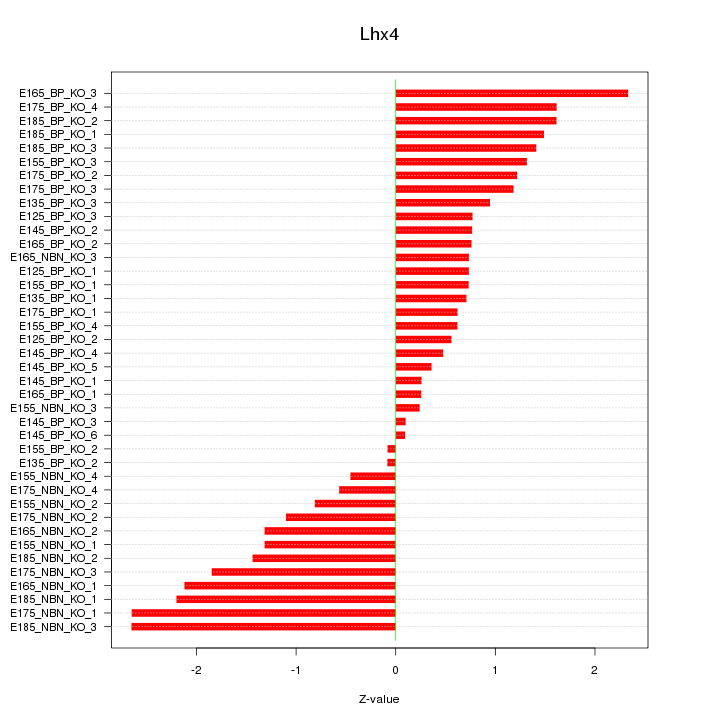
Motif ID: Lhx4
Z-value: 1.232
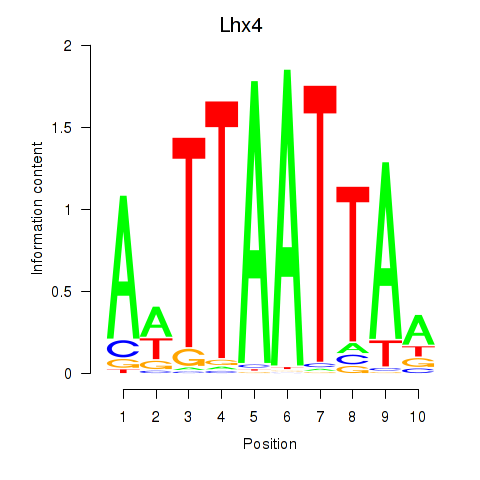
Transcription factors associated with Lhx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lhx4 | ENSMUSG00000026468.8 | Lhx4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 27.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.9 | 11.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.2 | 8.6 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.1 | 6.4 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 2.0 | 6.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.8 | 5.4 | GO:0060423 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 1.6 | 7.9 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 1.5 | 23.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.4 | 5.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.1 | 7.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.0 | 4.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.0 | 3.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.8 | 2.5 | GO:2000564 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.8 | 7.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.7 | 2.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.7 | 3.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.6 | 3.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.6 | 1.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 10.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.5 | 1.8 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.4 | 4.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 2.0 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 3.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 1.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.4 | 1.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 | 4.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.3 | 2.7 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 1.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 3.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 0.6 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.3 | 14.1 | GO:0030901 | midbrain development(GO:0030901) |
| 0.3 | 1.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.3 | 1.3 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.3 | 3.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 4.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.7 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 1.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.2 | 1.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 0.9 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.2 | 0.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.8 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) axonogenesis involved in innervation(GO:0060385) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.4 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 3.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.8 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 2.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) protein oxidation(GO:0018158) |
| 0.1 | 1.2 | GO:0036159 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.1 | 2.6 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.1 | 4.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.6 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 4.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 5.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 2.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 4.3 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.7 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 4.1 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 1.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.9 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.7 | 2.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.7 | 5.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.6 | 1.9 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.6 | 3.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 7.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 2.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 1.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 1.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 6.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 4.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.9 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 5.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 11.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 7.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 4.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 2.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 3.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 2.0 | 6.0 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.6 | 23.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.4 | 4.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.3 | 3.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 2.8 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.9 | 2.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.7 | 3.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 39.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.5 | 2.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 1.8 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.4 | 1.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.3 | 1.3 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.3 | 4.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 4.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 2.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 2.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 1.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 3.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.6 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 1.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 3.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 5.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 4.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 10.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 5.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 2.6 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 17.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 5.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 4.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 5.5 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 3.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 3.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 8.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.3 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 4.1 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.2 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.3 | 11.5 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 5.4 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 8.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.9 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.0 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.1 | 5.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.7 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.1 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.1 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 4.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 1.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.6 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 2.0 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.6 | 2.8 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 6.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.9 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.7 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 6.2 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 4.2 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.1 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.9 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.0 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.9 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 4.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.5 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.2 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.7 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 2.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.6 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.7 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |