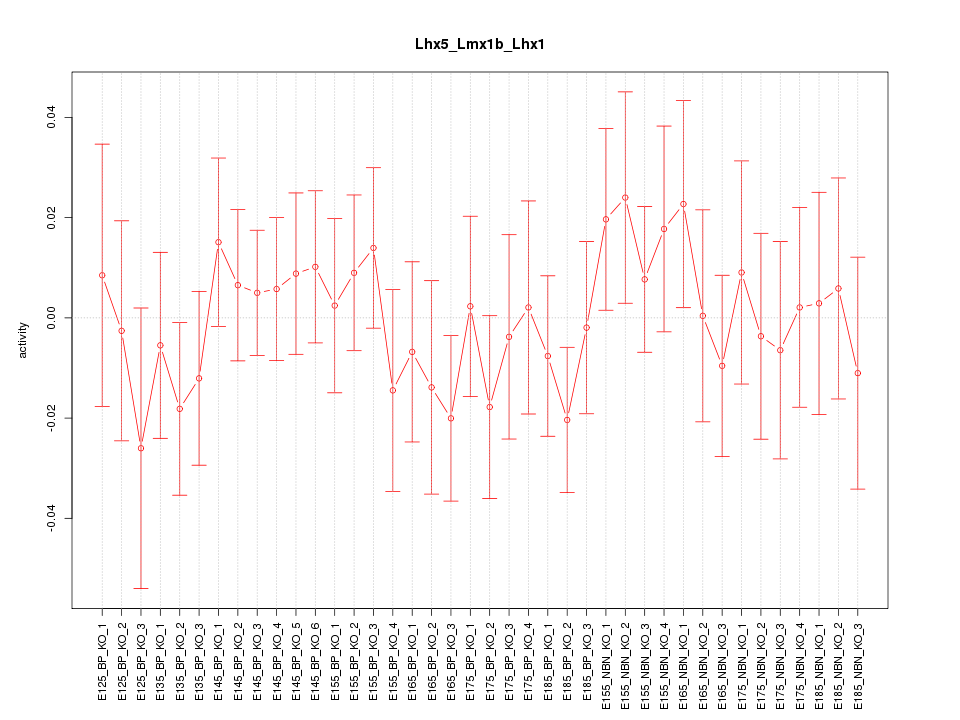
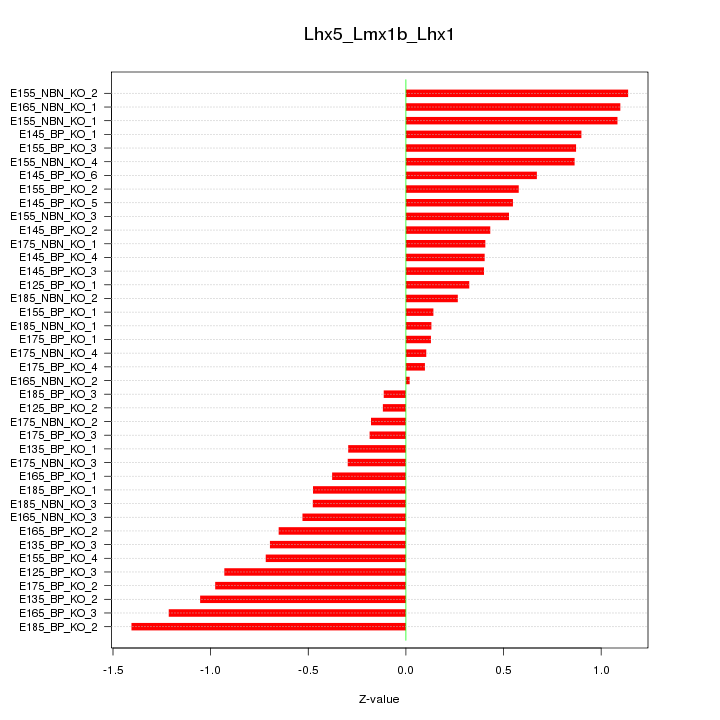
Motif ID: Lhx5_Lmx1b_Lhx1
Z-value: 0.656
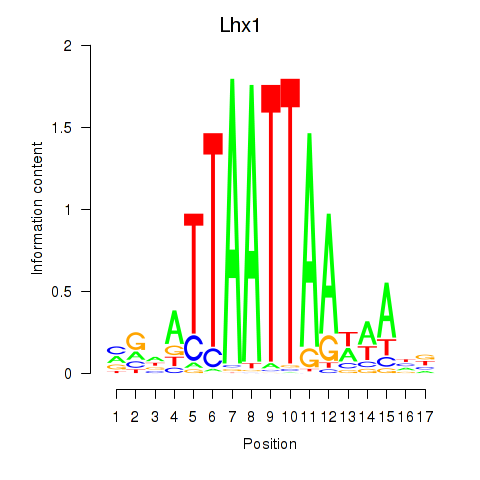
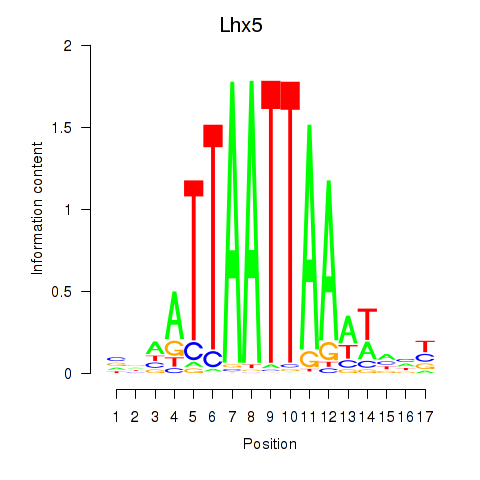
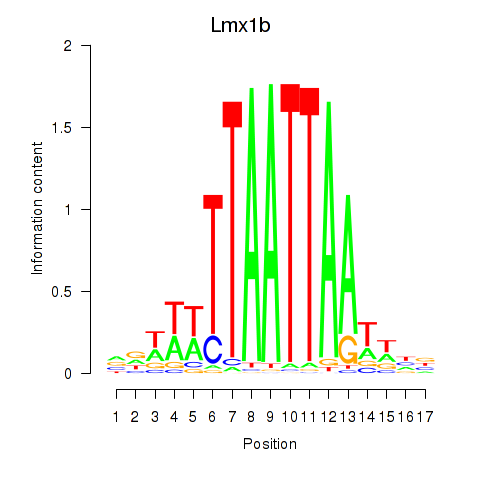
Transcription factors associated with Lhx5_Lmx1b_Lhx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lhx1 | ENSMUSG00000018698.9 | Lhx1 |
| Lhx5 | ENSMUSG00000029595.7 | Lhx5 |
| Lmx1b | ENSMUSG00000038765.7 | Lmx1b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx5 | mm10_v2_chr5_+_120431770_120431905 | -0.39 | 1.2e-02 | Click! |
| Lhx1 | mm10_v2_chr11_-_84525514_84525542 | -0.29 | 7.5e-02 | Click! |
| Lmx1b | mm10_v2_chr2_-_33640480_33640511 | -0.24 | 1.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.9 | 4.6 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.6 | 4.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.5 | 1.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 5.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 1.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.3 | 1.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.7 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 2.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.4 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.2 | 0.8 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.2 | GO:0039533 | MDA-5 signaling pathway(GO:0039530) regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.1 | 0.2 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 | 0.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 2.4 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 2.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.4 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.0 | 0.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 1.5 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 4.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 0.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 6.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.5 | 2.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 1.5 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.4 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 4.1 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.0 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.2 | 1.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 5.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 4.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 2.6 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.6 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.7 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.4 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.0 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.4 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.6 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.6 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 4.1 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 4.1 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 3.7 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |