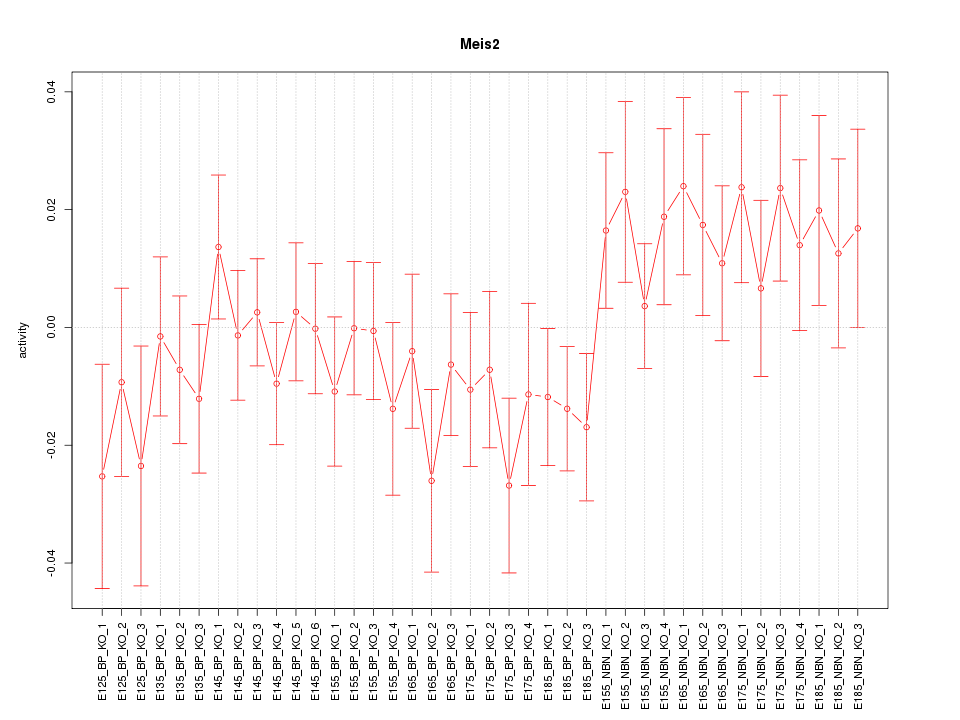
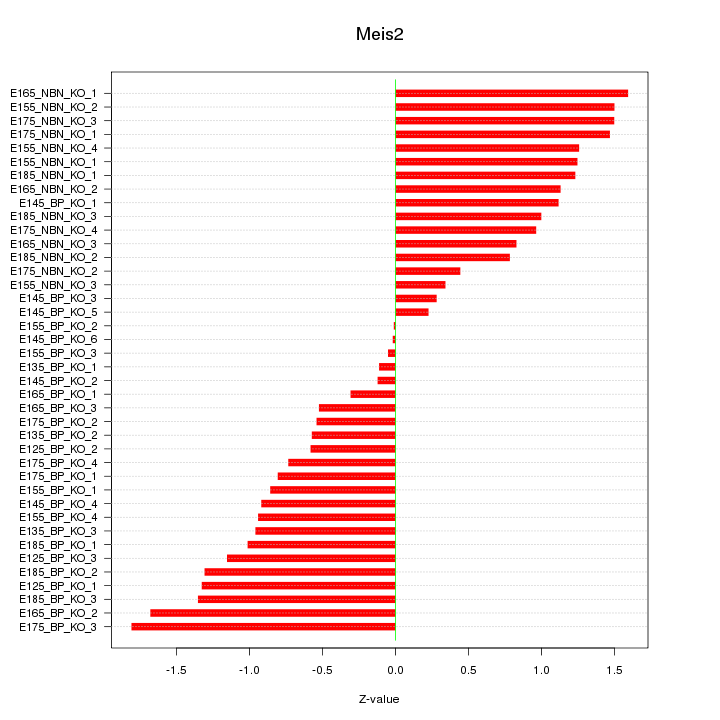
Motif ID: Meis2
Z-value: 0.997
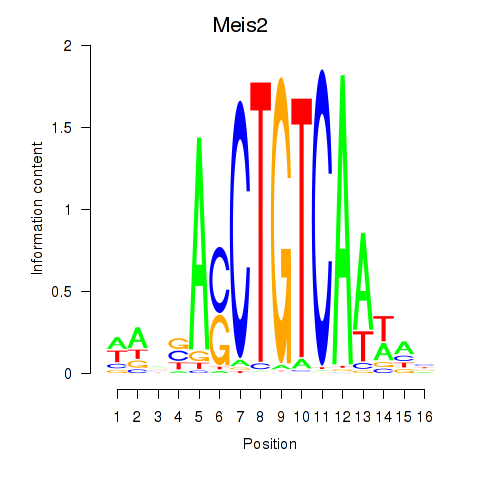
Transcription factors associated with Meis2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Meis2 | ENSMUSG00000027210.14 | Meis2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis2 | mm10_v2_chr2_-_116065047_116065133 | -0.31 | 5.0e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.6 | 7.8 | GO:1903587 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.5 | 4.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.4 | 4.3 | GO:0044413 | regulation of lung blood pressure(GO:0014916) evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 1.1 | 3.3 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.1 | 4.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.9 | 14.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.9 | 3.7 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.7 | 2.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.7 | 2.6 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.6 | 4.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.5 | 1.6 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.5 | 3.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.5 | 2.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.5 | 2.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 3.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.4 | 2.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 3.4 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 | 0.8 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.4 | 1.2 | GO:0061744 | motor behavior(GO:0061744) |
| 0.4 | 1.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 | 4.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 2.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 4.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.3 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 1.0 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.3 | 2.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 2.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.3 | 1.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 3.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 1.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 1.8 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.3 | 0.8 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 23.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.3 | 0.8 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.3 | 2.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 0.8 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 1.4 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 0.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 1.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.6 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 1.0 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 2.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 1.0 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 1.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 0.5 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.2 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 6.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.2 | GO:0039519 | modulation by virus of host autophagy(GO:0039519) |
| 0.2 | 0.8 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 0.5 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 2.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.4 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.4 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.4 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.9 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 3.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.1 | 0.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 2.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.4 | GO:0048296 | negative regulation of isotype switching(GO:0045829) isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 0.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 2.6 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.7 | GO:0007567 | parturition(GO:0007567) ductus arteriosus closure(GO:0097070) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 0.8 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.1 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.4 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 2.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.6 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.1 | 0.6 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 0.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 2.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 2.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 3.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 2.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.6 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 2.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.9 | 4.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.9 | 4.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.7 | 2.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.7 | 2.2 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.3 | 2.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 4.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 1.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 2.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 4.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 3.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 1.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 4.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 3.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.5 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.2 | 23.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 3.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.8 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.1 | 1.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 3.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 14.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.1 | 3.3 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.9 | 3.8 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.9 | 2.6 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 4.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.9 | 4.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.7 | 8.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.7 | 13.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 3.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 2.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.4 | 2.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 2.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 2.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 2.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 1.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 1.8 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.3 | 1.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 4.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 0.8 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 0.8 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 1.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.9 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.8 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 0.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 2.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 5.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 3.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 0.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 2.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.9 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 1.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 4.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.7 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 5.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 7.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 4.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 21.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 5.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 1.3 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 4.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.4 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.5 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.1 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.8 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.7 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.0 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.0 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 23.8 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 10.9 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 19.9 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 2.4 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 4.3 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 6.0 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 2.1 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.4 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 2.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.8 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.0 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.9 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.7 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 0.4 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.9 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.4 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.0 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |