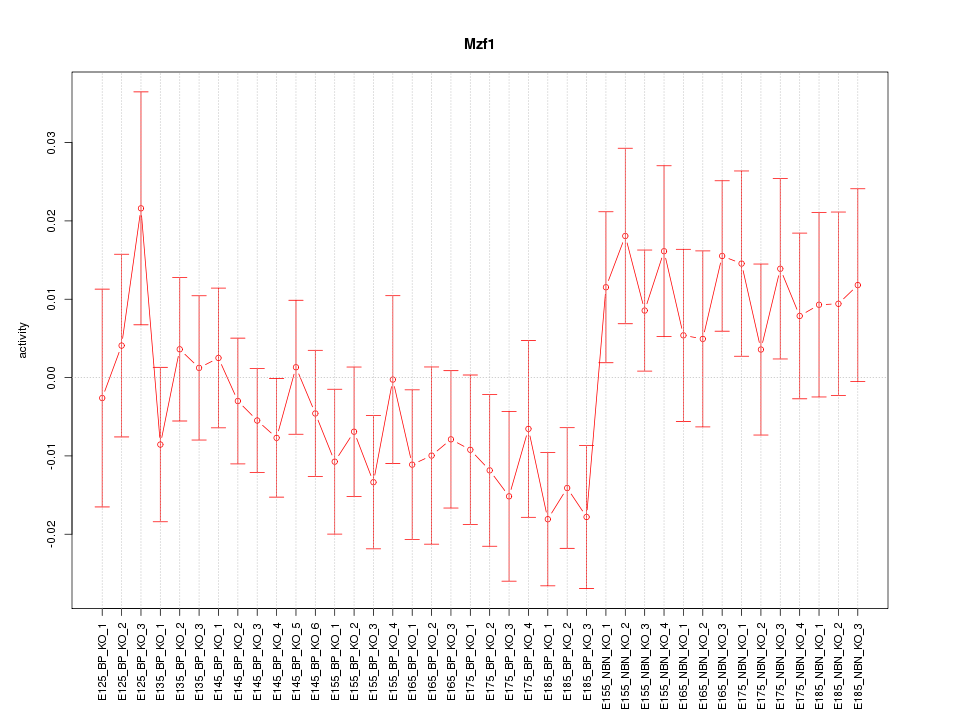
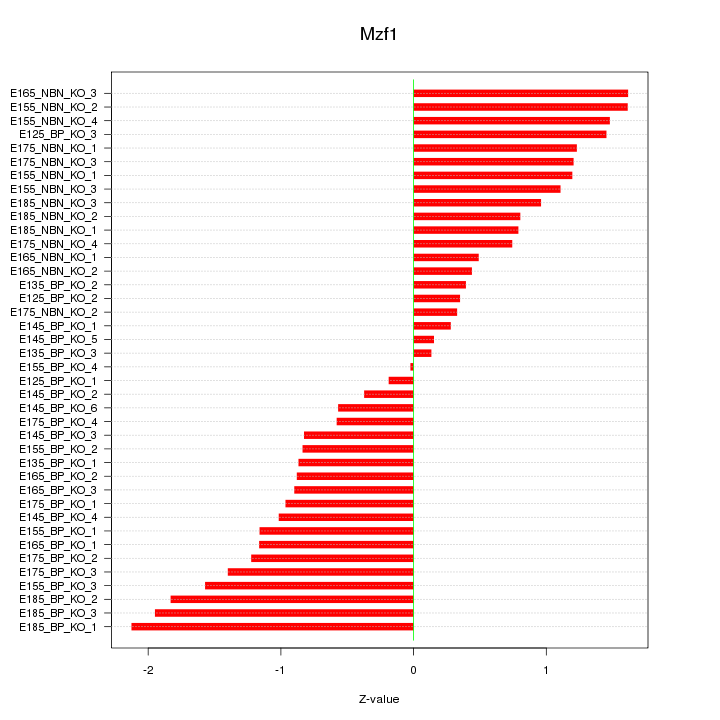
Motif ID: Mzf1
Z-value: 1.068
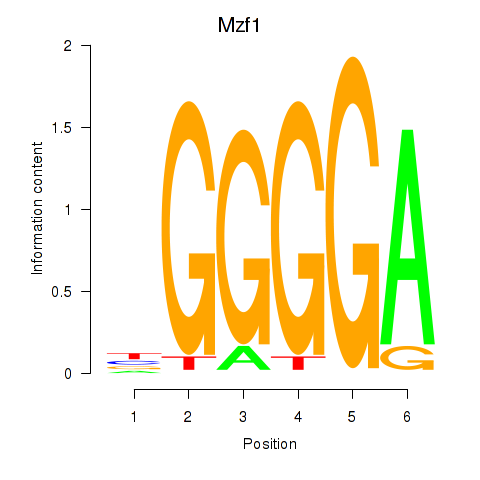
Transcription factors associated with Mzf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mzf1 | ENSMUSG00000030380.10 | Mzf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mzf1 | mm10_v2_chr7_-_13054665_13054764 | 0.42 | 6.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 2.0 | 6.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.5 | 4.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 1.2 | 4.7 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.9 | 2.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.9 | 4.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.9 | 2.6 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.8 | 2.5 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.8 | 3.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.8 | 7.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 3.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.7 | 2.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.7 | 9.0 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.7 | 3.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 4.6 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.7 | 2.0 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 3.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.6 | 1.9 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.6 | 1.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.6 | 1.7 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.6 | 1.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.6 | 4.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.5 | 2.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.5 | 1.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 0.5 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.5 | 1.6 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.5 | 2.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 4.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.5 | 3.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 1.5 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.5 | 2.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 1.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 1.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.5 | 1.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.5 | 8.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.4 | 2.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 1.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 | 1.3 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.4 | 1.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 2.5 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.4 | 1.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 2.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 1.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.4 | 1.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.4 | 1.9 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 | 1.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 3.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 3.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 2.6 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.4 | 1.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.4 | 2.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.3 | 1.4 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.3 | 0.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 | 1.3 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.3 | 1.6 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 0.3 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.3 | 0.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.3 | 0.9 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 | 0.9 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.3 | 0.9 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 1.2 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.3 | 1.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 0.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.3 | 3.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.3 | 1.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 2.7 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 0.8 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) cholangiocyte proliferation(GO:1990705) |
| 0.3 | 0.8 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.3 | 1.3 | GO:1903598 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 1.9 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 0.8 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 1.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 1.0 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 3.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 4.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 1.4 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 0.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 1.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 1.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.2 | 1.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 0.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 1.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 3.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 0.8 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 0.8 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 0.8 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 3.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 1.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 0.6 | GO:0070889 | platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.2 | 0.9 | GO:0032490 | detection of molecule of bacterial origin(GO:0032490) |
| 0.2 | 2.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 3.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 0.9 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 | 1.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.9 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 2.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 1.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.2 | GO:0071637 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.2 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.3 | GO:0010985 | negative regulation of lipoprotein particle clearance(GO:0010985) negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 0.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.5 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 | 1.3 | GO:0048265 | response to pain(GO:0048265) |
| 0.2 | 1.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 2.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.7 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 0.8 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 0.6 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 1.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 2.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 0.6 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.2 | 2.0 | GO:0043252 | prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.4 | GO:0098907 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 10.0 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 1.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 1.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 2.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.4 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.7 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.3 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.1 | 0.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 2.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.5 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 3.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) protein auto-ADP-ribosylation(GO:0070213) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0061419 | coronary vein morphogenesis(GO:0003169) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 2.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.1 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 0.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 1.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 1.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 5.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 2.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.7 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.3 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 0.5 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 2.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.3 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.1 | 0.4 | GO:0040032 | negative regulation of renal sodium excretion(GO:0035814) post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.6 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.1 | 0.2 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 0.4 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.5 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.9 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.2 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.7 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 2.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0002922 | positive regulation of humoral immune response(GO:0002922) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.5 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 1.5 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.2 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.4 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 0.0 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 2.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 1.6 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.0 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 4.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 1.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 2.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:1900364 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.0 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.2 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.7 | 2.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.6 | 2.5 | GO:0090537 | CERF complex(GO:0090537) |
| 0.6 | 9.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.6 | 0.6 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.5 | 2.7 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.5 | 2.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.5 | 10.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 4.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 8.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 11.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 2.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 1.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 3.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 1.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 5.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 3.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 0.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 0.9 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.3 | 0.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 11.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 2.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 16.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 6.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 2.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 2.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 3.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.2 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 2.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 24.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 2.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.9 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0097059 | ciliary neurotrophic factor receptor complex(GO:0070110) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.7 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 3.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0070905 | serine binding(GO:0070905) |
| 1.3 | 1.3 | GO:0048185 | activin binding(GO:0048185) |
| 1.2 | 7.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.1 | 4.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.9 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.9 | 2.6 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.8 | 6.7 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.8 | 2.5 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.7 | 3.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.7 | 4.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 1.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 2.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.5 | 2.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 4.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.5 | 7.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.5 | 3.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 2.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 4.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 1.9 | GO:0070891 | peptidoglycan binding(GO:0042834) lipoteichoic acid binding(GO:0070891) |
| 0.5 | 1.8 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.4 | 2.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 1.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.4 | 1.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 1.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 1.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 1.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 1.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 1.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 3.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 5.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 2.0 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 0.9 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 4.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 4.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 11.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 0.8 | GO:0034190 | very-low-density lipoprotein particle binding(GO:0034189) apolipoprotein receptor binding(GO:0034190) |
| 0.3 | 0.8 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.3 | 1.1 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.3 | 2.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 1.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 1.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 2.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 3.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.2 | 0.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.6 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 0.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 1.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 0.8 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.2 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.1 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.1 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 2.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 3.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 4.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 4.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.6 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 5.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 3.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 4.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.9 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 6.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 6.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 2.4 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 2.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 1.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 4.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 10.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.3 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 2.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 2.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.8 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 5.2 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.3 | 3.0 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.3 | 0.8 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 8.8 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 6.6 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.2 | 3.4 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.8 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.5 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 4.5 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.8 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.0 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.7 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.6 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.9 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 0.2 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.6 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.5 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.7 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.1 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.4 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 6.5 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 1.4 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID_MYC_PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.8 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.6 | 7.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 4.5 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.2 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 2.0 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 10.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 5.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.9 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 5.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 4.9 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 6.5 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.7 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.3 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.6 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.2 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.8 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.5 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.2 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 1.3 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.1 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.4 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.2 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.5 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.3 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.7 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.2 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.4 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.7 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.9 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.0 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.6 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.8 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.6 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.5 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 4.0 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME_CYCLIN_E_ASSOCIATED_EVENTS_DURING_G1_S_TRANSITION_ | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.8 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.1 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |