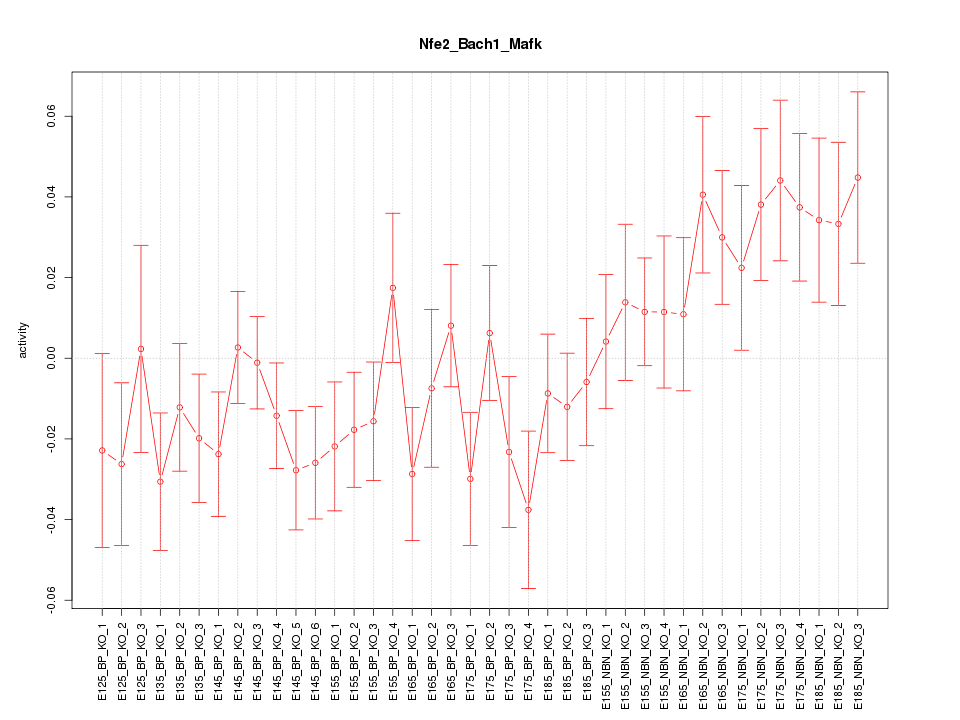
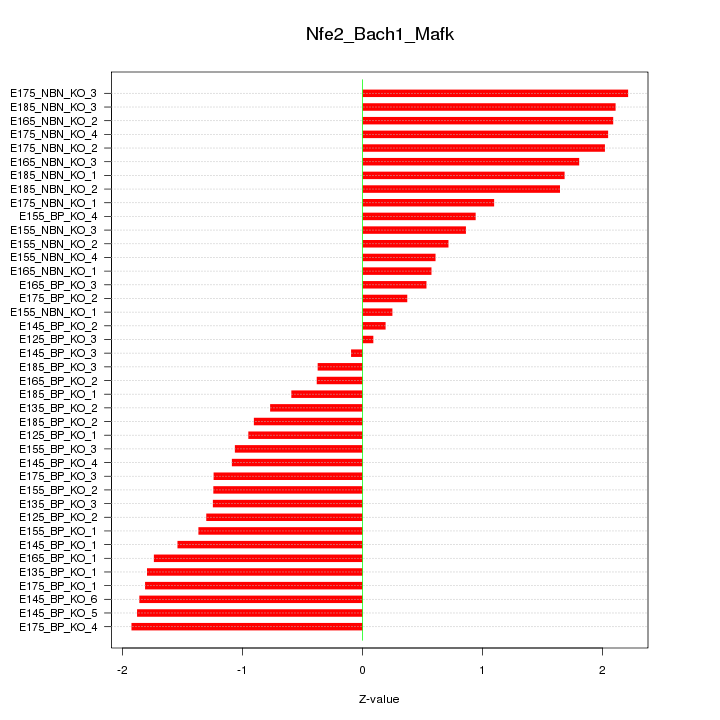
Motif ID: Nfe2_Bach1_Mafk
Z-value: 1.337
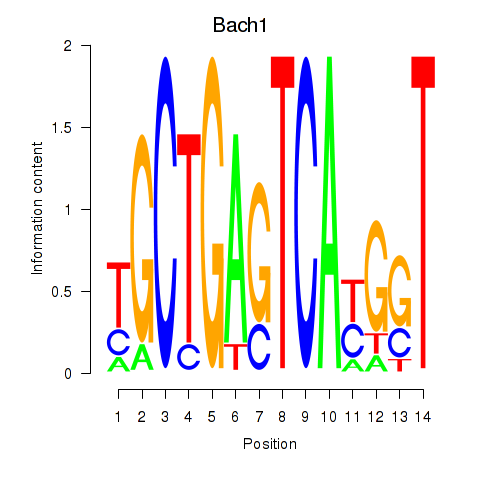
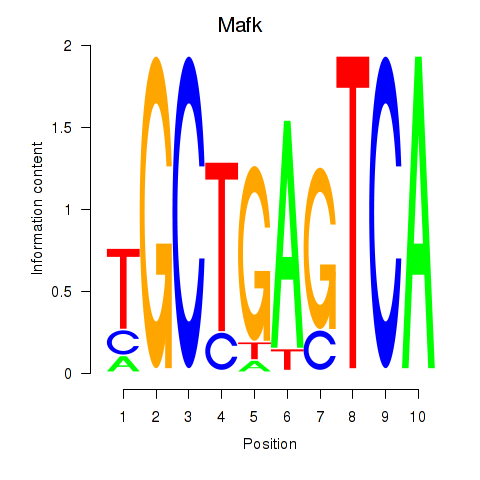
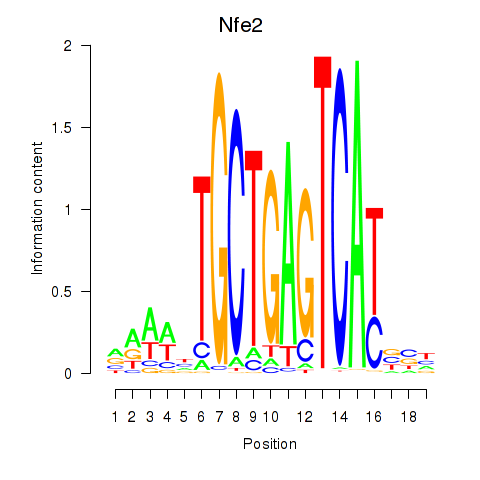
Transcription factors associated with Nfe2_Bach1_Mafk:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bach1 | ENSMUSG00000025612.5 | Bach1 |
| Mafk | ENSMUSG00000018143.4 | Mafk |
| Nfe2 | ENSMUSG00000058794.6 | Nfe2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafk | mm10_v2_chr5_+_139791513_139791539 | 0.81 | 3.6e-10 | Click! |
| Bach1 | mm10_v2_chr16_+_87698904_87698959 | -0.55 | 2.1e-04 | Click! |
| Nfe2 | mm10_v2_chr15_-_103251465_103251705 | -0.35 | 2.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 36.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 2.4 | 12.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.8 | 9.0 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 1.6 | 4.8 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 1.6 | 4.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.6 | 4.7 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.3 | 5.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.3 | 6.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 1.2 | 3.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 1.1 | 2.3 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 1.1 | 7.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.9 | 3.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.8 | 2.5 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.8 | 2.4 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.8 | 4.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.8 | 5.4 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.7 | 6.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.7 | 9.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.6 | 3.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.6 | 5.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 2.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.5 | 6.6 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.5 | 4.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 2.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.5 | 2.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.5 | 3.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.4 | 16.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.4 | 1.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 2.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 1.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 1.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.4 | 2.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 1.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 2.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 0.9 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.3 | 2.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 6.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 2.2 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.3 | 2.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 1.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.9 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 2.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 3.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 1.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.8 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.2 | 1.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.2 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 0.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 1.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.4 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.2 | 4.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 0.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 4.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 2.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.8 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.2 | 0.6 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.5 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 1.5 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 10.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.9 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.4 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 0.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.5 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.2 | GO:0035482 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.1 | 2.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 0.9 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.5 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 1.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.4 | GO:0072224 | metanephric glomerulus development(GO:0072224) |
| 0.1 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 3.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.7 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 2.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.5 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 3.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.3 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 | 0.4 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 1.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 5.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.9 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.3 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 4.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 3.1 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 1.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 0.9 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 36.5 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.4 | 4.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.9 | 10.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.7 | 2.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 2.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 5.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.5 | 1.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 6.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 3.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 4.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 4.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 4.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 4.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 0.7 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.2 | 13.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 2.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 4.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 5.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.5 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 1.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 2.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 7.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 7.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 3.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.0 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 6.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 5.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0038201 | TORC1 complex(GO:0031931) TOR complex(GO:0038201) |
| 0.0 | 0.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 4.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.4 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.8 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 2.1 | 36.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.9 | 7.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.4 | 5.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.0 | 5.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 4.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.9 | 10.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 4.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 4.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 2.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 17.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.5 | 3.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 1.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 6.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 5.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 2.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 3.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 1.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 1.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 6.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 0.8 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.3 | 0.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 1.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 2.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 2.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 2.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 3.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 5.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 3.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 4.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.7 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 4.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.3 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 2.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 3.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 3.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 3.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 9.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.0 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 10.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.3 | 9.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 3.9 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.2 | 7.6 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 9.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.5 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.1 | 7.6 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.2 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.6 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 8.8 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.4 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 4.4 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.6 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 1.6 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.0 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 6.0 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.7 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.2 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.2 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.9 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID_ATR_PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.9 | 4.3 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.8 | 11.8 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.7 | 6.9 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 12.9 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 44.3 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.4 | 2.8 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 4.7 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 2.5 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 5.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.2 | 5.0 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.1 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 3.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.7 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 7.7 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 4.6 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 2.1 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 9.1 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.9 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.4 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.2 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 5.8 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.3 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 2.5 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |