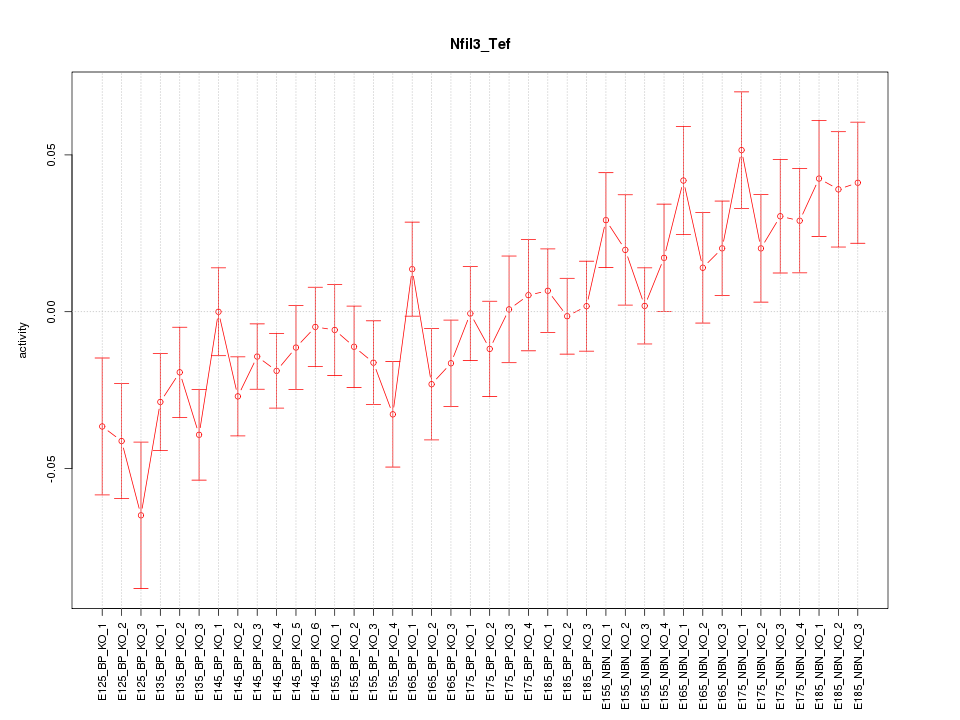
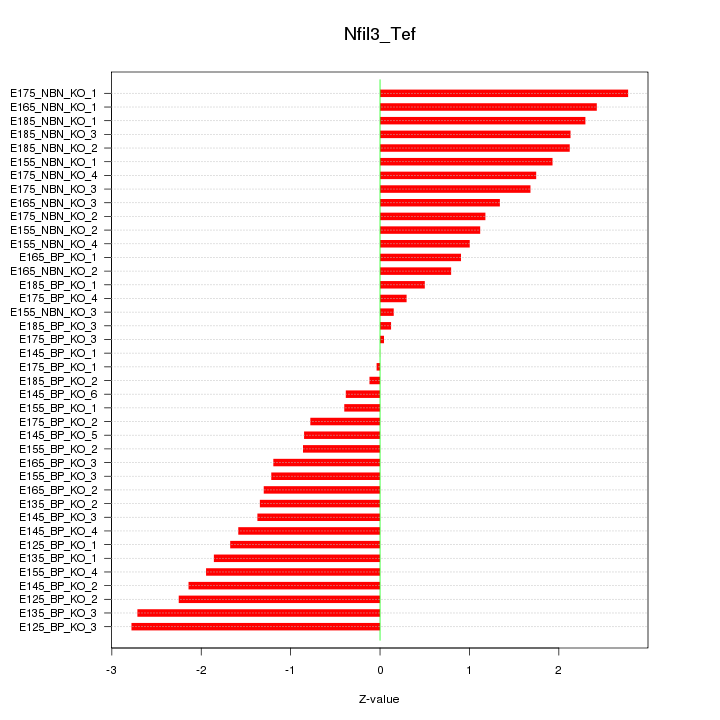
Motif ID: Nfil3_Tef
Z-value: 1.523
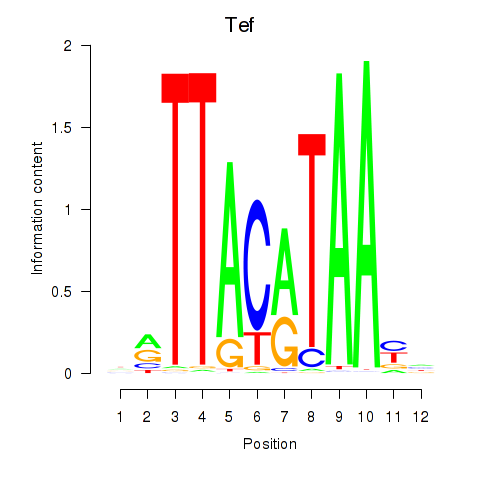
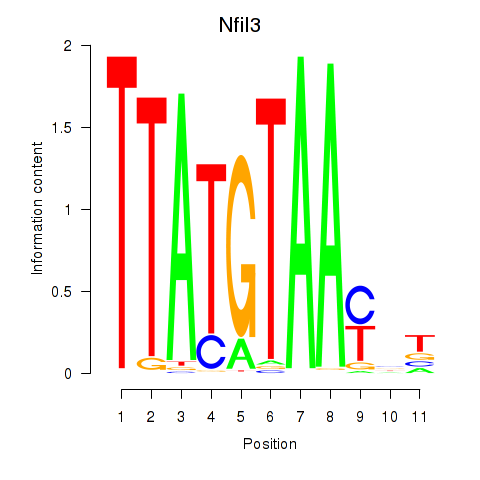
Transcription factors associated with Nfil3_Tef:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfil3 | ENSMUSG00000056749.7 | Nfil3 |
| Tef | ENSMUSG00000022389.8 | Tef |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfil3 | mm10_v2_chr13_-_52981027_52981083 | 0.54 | 3.5e-04 | Click! |
| Tef | mm10_v2_chr15_+_81811414_81811491 | 0.27 | 8.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 35.9 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 4.3 | 25.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 3.3 | 22.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 3.0 | 12.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.9 | 8.7 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 2.3 | 7.0 | GO:0099548 | drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 2.0 | 5.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 1.9 | 5.7 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.7 | 11.9 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 1.7 | 6.8 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.7 | 6.7 | GO:2000484 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) positive regulation of interleukin-8 secretion(GO:2000484) |
| 1.6 | 8.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 1.6 | 4.9 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.5 | 10.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.2 | 12.0 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 1.2 | 2.3 | GO:0060558 | regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 1.1 | 3.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.1 | 6.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.1 | 8.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.0 | 5.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.8 | 2.3 | GO:0061744 | motor behavior(GO:0061744) |
| 0.8 | 2.3 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.7 | 3.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.7 | 4.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 5.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.7 | 3.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.6 | 1.8 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.6 | 1.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 1.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 4.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 1.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.5 | 1.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 3.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 4.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.5 | 4.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 4.2 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 1.4 | GO:2001180 | regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.4 | 2.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.4 | 1.9 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 3.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.4 | 5.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 3.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 0.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.3 | 1.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 | 2.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 1.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 1.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.7 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 5.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 1.1 | GO:0006048 | glucosamine metabolic process(GO:0006041) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 1.7 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 0.6 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 1.4 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.2 | 5.7 | GO:0071625 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) vocalization behavior(GO:0071625) |
| 0.2 | 1.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 4.8 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.2 | 1.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.9 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 3.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.4 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.4 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 1.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 4.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 1.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 5.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 1.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 8.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.9 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 4.1 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.4 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 35.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 2.7 | 8.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 2.0 | 8.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.3 | 5.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.1 | 6.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 2.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.6 | 7.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.6 | 5.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 1.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 1.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 3.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 5.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 5.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 4.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 5.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 3.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 6.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 6.4 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) postsynaptic specialization(GO:0099572) |
| 0.2 | 1.6 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 0.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 7.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.1 | 5.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.4 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 5.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 14.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 5.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 3.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 6.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 51.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 2.5 | 7.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 2.2 | 6.7 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 2.0 | 8.1 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 1.9 | 5.7 | GO:0031403 | lithium ion binding(GO:0031403) |
| 1.7 | 6.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.6 | 9.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.5 | 35.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.4 | 8.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.4 | 8.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.4 | 4.1 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 1.2 | 4.9 | GO:1903135 | cupric ion binding(GO:1903135) |
| 1.2 | 3.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.1 | 4.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.0 | 5.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.9 | 3.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.8 | 3.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.8 | 2.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.8 | 2.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.6 | 1.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.6 | 4.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 1.8 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 2.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.6 | 1.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.5 | 8.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 3.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 2.3 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.5 | 1.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.4 | 1.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.4 | 1.7 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.4 | 1.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 3.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 5.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 5.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 5.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 0.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 5.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 2.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 0.5 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 4.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 3.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 8.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 2.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 6.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 16.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 2.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 8.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 16.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 4.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 9.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 1.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.4 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 8.7 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.4 | 35.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 2.3 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 6.7 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.2 | 8.2 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.2 | 5.1 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.2 | 10.2 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.2 | 8.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.2 | 5.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.2 | 8.7 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 5.9 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.8 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.3 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 7.3 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 0.7 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 4.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 1.9 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.9 | 4.3 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.7 | 4.9 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.7 | 25.7 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.6 | 5.8 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 6.7 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.6 | 5.7 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 10.2 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 35.9 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 7.6 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 7.1 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 11.2 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 8.7 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 8.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 8.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.3 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 6.1 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 7.5 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 5.0 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.0 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.6 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.3 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.2 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.7 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 0.5 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.3 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.5 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.9 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.3 | REACTOME_MRNA_PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.0 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |