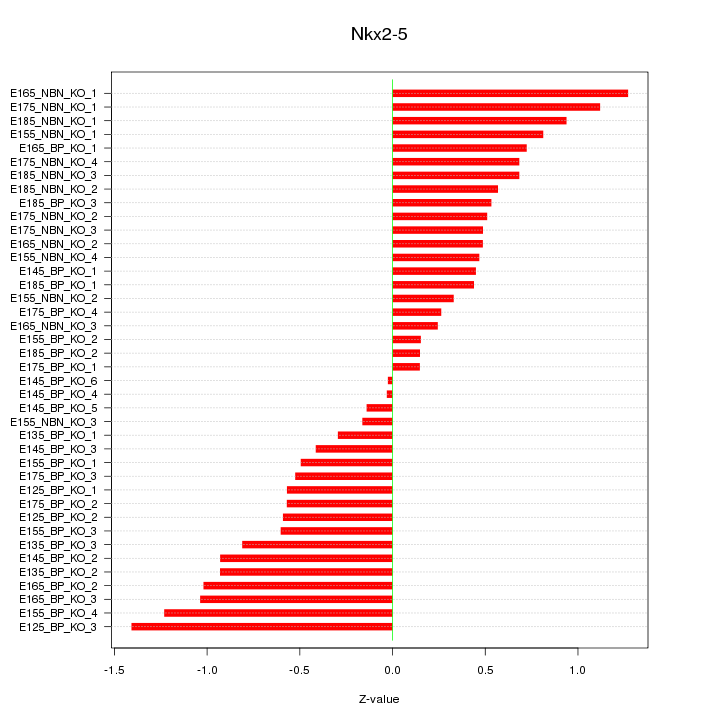
Motif ID: Nkx2-5
Z-value: 0.677

Transcription factors associated with Nkx2-5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nkx2-5 | ENSMUSG00000015579.4 | Nkx2-5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.1 | 3.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.6 | 1.7 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) regulation of microglial cell activation(GO:1903978) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 3.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 5.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) cellular response to cholesterol(GO:0071397) |
| 0.5 | 2.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 2.4 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.4 | 1.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 3.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.3 | 1.6 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.3 | 2.4 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.2 | 0.9 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 4.6 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.2 | 0.7 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.1 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.1 | 0.9 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 1.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.8 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.1 | 1.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 2.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.6 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 1.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 3.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 3.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 3.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 5.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.6 | 1.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.5 | 2.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 3.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 2.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 5.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.9 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 5.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.2 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 7.1 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 2.4 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.4 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 4.2 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.7 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 4.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.4 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.7 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 1.7 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.8 | REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.8 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.9 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.2 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |