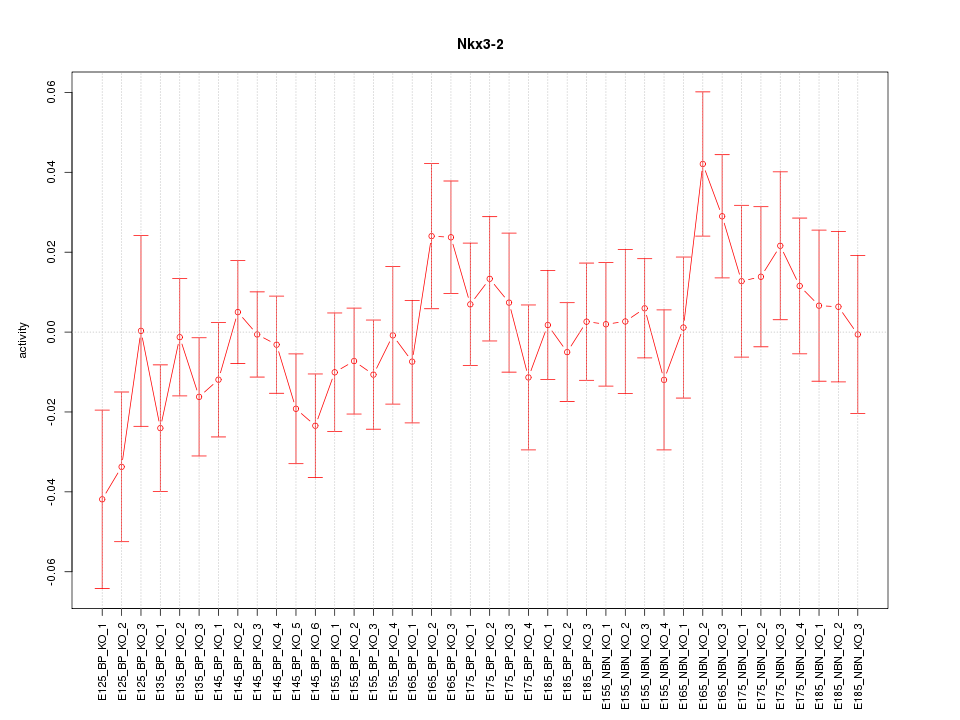
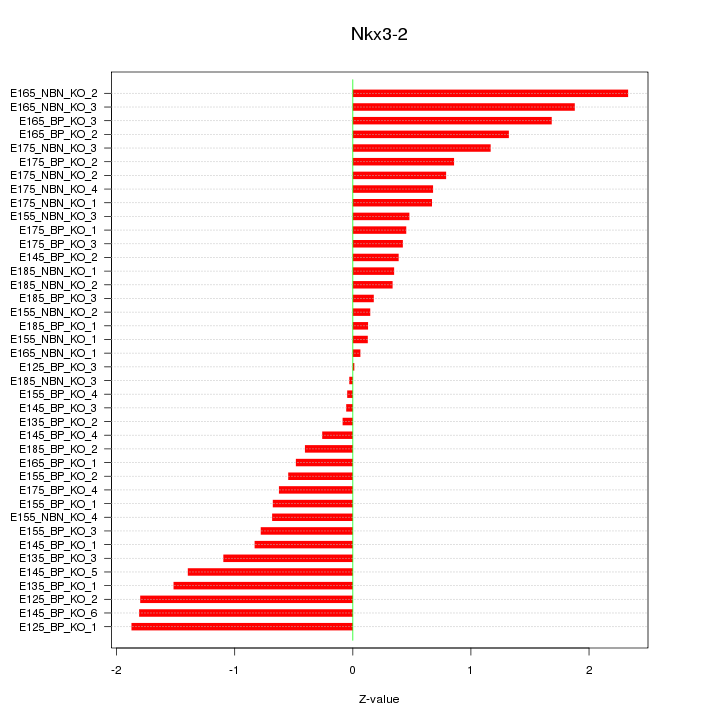
Motif ID: Nkx3-2
Z-value: 0.964

Transcription factors associated with Nkx3-2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nkx3-2 | ENSMUSG00000049691.7 | Nkx3-2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.9 | 2.6 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.8 | 3.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.7 | 3.7 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.6 | 2.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 1.9 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.6 | 2.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 2.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.5 | 5.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.5 | 1.5 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 1.4 | GO:0003289 | endocardial cushion fusion(GO:0003274) atrial septum primum morphogenesis(GO:0003289) |
| 0.5 | 1.8 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.4 | 2.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 4.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.4 | 2.5 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 0.8 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.4 | 1.6 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.4 | 1.2 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.4 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 1.0 | GO:2000319 | negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 1.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 1.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.3 | 1.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 3.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 7.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 2.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 1.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 0.9 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 1.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.3 | 0.8 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.3 | 1.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 1.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.5 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.2 | 1.5 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.2 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 2.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 1.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 0.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 | 4.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 6.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 1.3 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.2 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.5 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 1.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 2.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.8 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 0.5 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.8 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 1.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 2.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.5 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.8 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.8 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.7 | GO:0001963 | synaptic transmission, dopaminergic(GO:0001963) |
| 0.1 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 2.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.6 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 1.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.5 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 1.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.0 | 0.6 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.6 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 1.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 1.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 1.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 3.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.4 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 5.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 3.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 3.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 1.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 1.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 2.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 8.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 5.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 5.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 6.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.8 | 3.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 2.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.6 | 2.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.5 | 3.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 1.5 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.4 | 1.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 1.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 1.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 3.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 1.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 3.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 1.8 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.3 | 3.0 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.3 | 1.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.4 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.3 | 0.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.9 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.7 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.2 | 2.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.6 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.2 | 7.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.3 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.9 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 1.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 2.0 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0070492 | sialic acid binding(GO:0033691) oligosaccharide binding(GO:0070492) |
| 0.1 | 1.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 4.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 4.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.5 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.4 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 1.5 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.7 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.1 | 3.7 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 7.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.4 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.8 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.4 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 4.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 3.3 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.5 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 1.9 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.7 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.6 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.8 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.8 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.3 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 2.0 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.7 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.3 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.8 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.1 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.3 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.7 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.5 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |