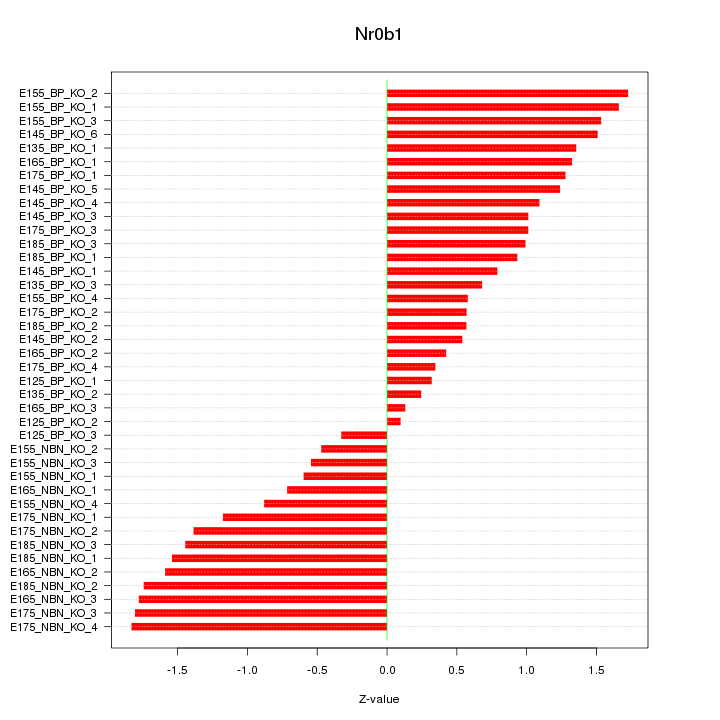
Motif ID: Nr0b1
Z-value: 1.123
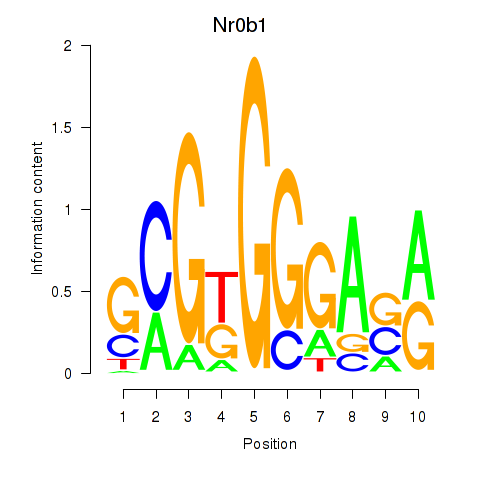
Transcription factors associated with Nr0b1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr0b1 | ENSMUSG00000025056.4 | Nr0b1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm10_v2_chrX_+_86191764_86191782 | 0.24 | 1.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 16.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.8 | 7.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.3 | 6.4 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) hard palate development(GO:0060022) |
| 1.2 | 4.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.8 | 10.5 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.7 | 2.9 | GO:0072236 | DCT cell differentiation(GO:0072069) metanephric loop of Henle development(GO:0072236) metanephric DCT cell differentiation(GO:0072240) |
| 0.7 | 2.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 3.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 | 1.6 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 3.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 3.3 | GO:2001225 | cellular hypotonic response(GO:0071476) regulation of chloride transport(GO:2001225) |
| 0.5 | 1.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 1.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) cell proliferation involved in heart valve development(GO:2000793) |
| 0.4 | 1.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 1.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 1.7 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.3 | 1.4 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.3 | 1.6 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 0.8 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.2 | 0.5 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.2 | 0.8 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.2 | 8.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 3.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 3.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 2.1 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 5.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.4 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) positive regulation of response to drug(GO:2001025) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.9 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 1.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 2.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 1.0 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.3 | GO:0034612 | response to tumor necrosis factor(GO:0034612) |
| 0.0 | 1.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 3.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.5 | 1.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 10.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.4 | 8.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 4.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 7.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 6.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.7 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 1.2 | 8.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.0 | 4.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.9 | 3.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.8 | 3.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 4.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.5 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.3 | 6.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 4.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 3.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 5.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 10.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 4.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 7.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 11.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.2 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.2 | 1.6 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 8.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.2 | 10.2 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.1 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 1.4 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.4 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.9 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 1.1 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.1 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.3 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.2 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID_REELIN_PATHWAY | Reelin signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.4 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 10.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.1 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 1.9 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 5.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.3 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.7 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 6.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.3 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.4 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |