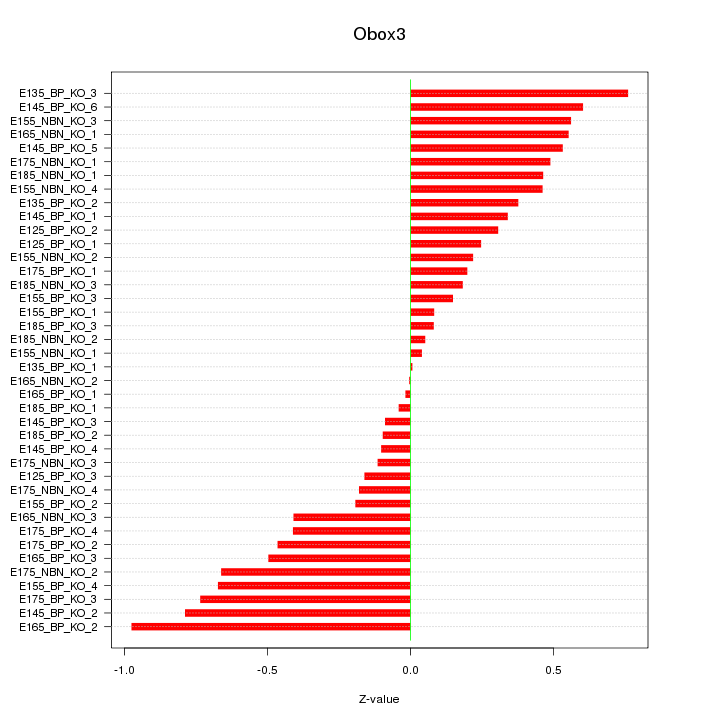
Motif ID: Obox3
Z-value: 0.419
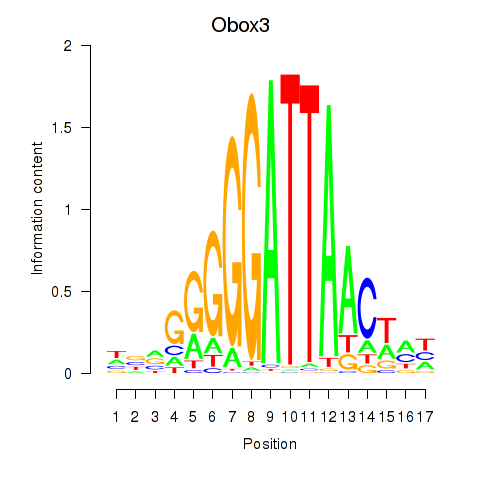
Transcription factors associated with Obox3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Obox3 | ENSMUSG00000066772.6 | Obox3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox3 | mm10_v2_chr7_-_15627876_15627876 | 0.01 | 9.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.5 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 3.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 1.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.1 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.5 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |