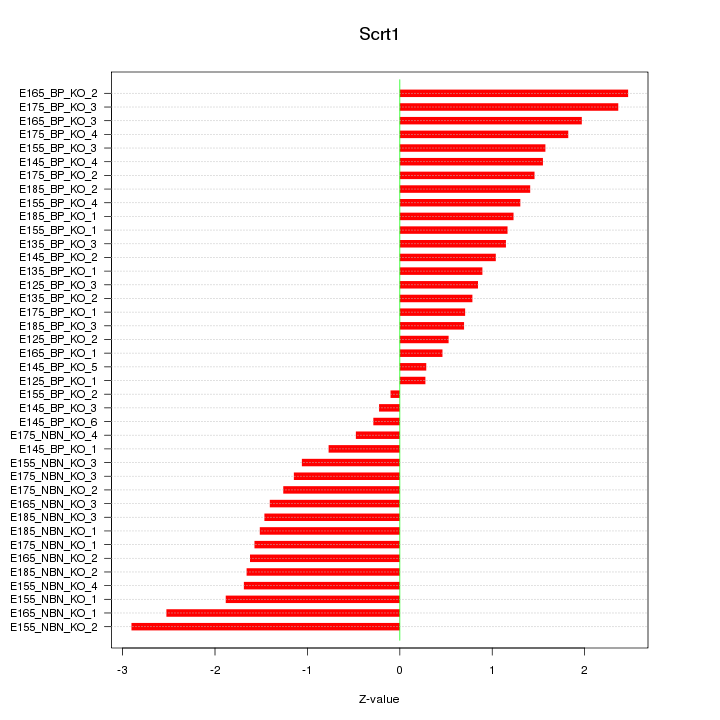
Motif ID: Scrt1
Z-value: 1.406

Transcription factors associated with Scrt1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Scrt1 | ENSMUSG00000048385.8 | Scrt1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt1 | mm10_v2_chr15_-_76521902_76522129 | -0.22 | 1.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 3.1 | 9.3 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 2.8 | 8.5 | GO:0061349 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 1.8 | 3.6 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 1.7 | 5.2 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 1.5 | 6.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 1.3 | 6.6 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.3 | 5.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.2 | 6.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.1 | 18.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.0 | 5.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.9 | 2.8 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 7.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.8 | 4.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.8 | 4.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.8 | 4.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.8 | 2.4 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.7 | 2.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.7 | 2.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.7 | 2.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.7 | 6.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 1.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.6 | 8.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 2.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.5 | 2.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.5 | 3.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.5 | 1.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) renal vesicle induction(GO:0072034) |
| 0.5 | 6.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.4 | 1.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 | 2.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 2.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 1.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 2.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 1.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.4 | 1.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 0.9 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.3 | 3.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 4.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.3 | 0.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 8.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.3 | 3.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 1.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 0.9 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.2 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 3.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 1.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 | 2.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 1.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.6 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 2.1 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.1 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 3.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 4.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 2.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 4.0 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 2.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 2.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.7 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 0.1 | 1.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 6.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 1.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.5 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 7.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.9 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.4 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.0 | 1.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 2.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 3.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.8 | GO:0071560 | cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.0 | 1.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 1.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.7 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 4.8 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 1.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.8 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 0.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 2.5 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.6 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.5 | 7.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 5.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 4.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 1.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 6.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 3.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 9.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 6.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 11.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 2.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 6.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 4.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 2.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.3 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 2.6 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 12.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 2.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 5.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 18.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 5.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0030677 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 5.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.3 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 6.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 11.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 1.3 | 5.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.3 | 5.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.8 | 6.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.8 | 2.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.7 | 3.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.7 | 2.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 9.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 1.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 1.4 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.4 | 2.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 4.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 10.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.4 | 1.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 7.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 5.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 2.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 2.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 9.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 1.9 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.3 | 11.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 1.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.3 | 6.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 10.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 4.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 2.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 0.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 3.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.4 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 3.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 4.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 6.6 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.5 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 1.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 5.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 8.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.6 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 1.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.1 | 3.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 4.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 29.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 18.2 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.4 | 7.5 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 15.1 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 4.3 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.6 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 2.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.4 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.2 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.3 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.2 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.9 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.8 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.1 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 1.9 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.7 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.3 | 6.5 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 6.2 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.2 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 5.8 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 1.3 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.9 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 8.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.6 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.9 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 2.8 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 7.5 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 0.6 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.9 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.4 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.9 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.2 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.8 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.1 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.9 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.5 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.5 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |